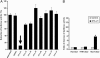A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells - PubMed (original) (raw)
A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells
Hui Song et al. Proc Natl Acad Sci U S A. 2005.
Abstract
This study focused on the screening of small-molecule inhibitors that target signal transducers and activators of transcription 3 (Stat3) in human breast carcinoma. The constitutive activation of Stat3 is frequently detected in human breast cancer cell lines as well as clinical breast cancer specimens and may play an important role in the oncogenesis of breast carcinoma. Activated Stat3 may participate in oncogenesis by stimulating cell proliferation, promoting tumor angiogenesis, and resisting apoptosis. Because a variety of human cancers are associated with constitutively active Stat3, Stat3 represents an attractive target for cancer therapy. In this study, of the nearly 429,000 compounds screened by virtual database screening, chemical samples of top 100 compounds identified as candidate small-molecule inhibitors of Stat3 were evaluated by using Stat3-dependent luciferase reporter as well as other cell-based assays. Through serial functional evaluation based on our established cell-based assays, one compound, termed STA-21, was identified as the best match for our selection criteria. Further investigation demonstrated that STA-21 inhibits Stat3 DNA binding activity, Stat3 dimerization, and Stat3-dependent luciferase activity. Moreover, STA-21 reduces the survival of breast carcinoma cells with constitutive Stat3 signaling but has minimal effect on the cells in which constitutive Stat3 signaling is absent. Together, these results demonstrate that STA-21 inhibits breast cancer cells that express constitutively active Stat3.
Figures
Fig. 1.
Schematic diagrams of modeling of structure-based virtual database screening. (A) SH2 domain dimerization interface of the STAT3β protein. The structure is based on Protein Data Bank entry 1BG1. Two SH2 domains are colored differently. The circled region indicates the target PTR-binding site used in our virtual screening study. (B) Predicted binding model of STA-21 to the STAT3β SH2 domain. STA-21 is rendered by a ball-and-stick model. The molecular surface of STAT3β SH2 domain is colored with the electrostatic potentials: red for most positively charged regions and blue for most negatively charged regions. (C) Specific hydrogen bonds formed between the STAT3β SH2 domain and STA-21. The binding model was predicted by
dock
. Only the residues that form hydrogen bonds with STA-21 are shown in explicit atomic models. (D) STA-21 structure. _A_-C were generated by using
sybyl
.
Fig. 2.
STA-21 inhibits Stat3-dependent luciferase activity in cancer cells. (A) The clone from Caov-3 carcinoma cells stably transfected with pLucTKS3 Stat3-dependent luciferase reporter was used for initial Sat3 small-molecule inhibitor screening. The cloned cells were treated with 20 μM STA-21 as well as other small-molecule compounds for 48 h, and then the cells were harvested for luciferase activity analysis. (B) The clones from MDA-MB-435s cells stably transfected with pLucTKS3 Stat3-dependent luciferase reporter or simian virus 40 (SV40) luciferase reporter were treated with 20 μM STA-21 for 48 h. Luciferase activity was measure by using a Promega luciferase kit according to the manufacturer's instructions. The results were based on the means and standard deviations from three separate experiments.
Fig. 3.
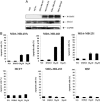
STA-21 inhibits Stat3 DNA binding activity and Stat3-regulated antiapoptotic factors. (A) MDA-MB-435s cell nucleus extract was incubated with 30 μM STA-21 for 30 min at room temperature and then incubated with r-[32P]ATP-labeled consensus binding sequence for 20 min at room temperature. The reaction mixtures were resolved on 8% polyacrylamide gel. (B) The lysates from MDA-MB-468 cells treated with indicated concentrations of STA-21 for 48 h were resolved on 10% SDS/PAGE, then immunoblotted with Abs as indicated.
Fig. 4.
STA-21 inhibits the survival of breast carcinoma cells with constitutive Stat3 signaling but not in cells without constitutive Stat3 signaling. (A) The phosphorylation of Stat3 at Tyr-705 in different cell lines. (B) The cell lines were treated with STA-21 at the indicated concentrations for 48 h, and then cells were harvested and analyzed for the SubG1 profile that indicated apoptotic cells on a FACScan Flow Cytometer (Becton Dickinson). The results were based on the means and standard deviations from three separate experiments.
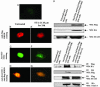
Fig. 5.
STA-21 inhibits Stat3 translocation and dimerization in breast carcinoma cells. The MDA-MB-435s cells cotransfected with pCMV-Stat3-Flag and pCMV-Stat3-HA plasmid were exposed to 20 μM STA-21 compound for 24 h, then fixed with 100% methanol. After the fixed cells were stained with anti-HA (rabbit, Santa Cruz Biotechnology) and/or anti-Flag (mouse, Sigma) Abs, secondary anti-rabbit IgG-FITC and/or anti-mouse IgG-rhodamine Abs were added. The cells were observed by using a fluorescence microscope. (A) Untransfected and untreated MDA-MB-435s cells. (B and E) Untreated (B) and STA-21-treated (E) cells cotransfected by pCMV-Stat3-Flag and pCMV-Stat3-HA plasmids were immunostained with anti-Flag IgG-rhodamine. (C and F) Untreated (C) and STA-21 treated (F) transfected cells were immunostained with anti-HA IgG-FITC. (D and G) The transfected cells were coimmunostained with both anti-HA IgG-FITC and anti-Flag IgG-rhodamine_. (D_) Untreated cells showed bright orange color. (G) STA-21 treated cells showed weak orange staining and separate green and red color. (Magnification: ×400.) (H) The MDA-MB-435s cells were cotransfected with pCMV-Stat3-Flag and pCMV-Stat3-HA plasmids and were exposed to 20 μM STA-21 compound for 24 h, and then cell lysates were immunoprecipitated with anti-HA or anti-Flag Abs, respectively, as described previously. Cells were resolved on 10% SDS/PAGE and then immunoblotted with anti-HA, anti-Flag or anti-Sat3 Abs as described in Materials and Methods.
Similar articles
- Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation.
Turkson J, Ryan D, Kim JS, Zhang Y, Chen Z, Haura E, Laudano A, Sebti S, Hamilton AD, Jove R. Turkson J, et al. J Biol Chem. 2001 Nov 30;276(48):45443-55. doi: 10.1074/jbc.M107527200. Epub 2001 Sep 28. J Biol Chem. 2001. PMID: 11579100 - Inhibition of constitutively active Stat3 suppresses growth of human ovarian and breast cancer cells.
Burke WM, Jin X, Lin HJ, Huang M, Liu R, Reynolds RK, Lin J. Burke WM, et al. Oncogene. 2001 Nov 29;20(55):7925-34. doi: 10.1038/sj.onc.1204990. Oncogene. 2001. PMID: 11753675 - Constitutive activation of Stat3 in human prostate tumors and cell lines: direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells.
Mora LB, Buettner R, Seigne J, Diaz J, Ahmad N, Garcia R, Bowman T, Falcone R, Fairclough R, Cantor A, Muro-Cacho C, Livingston S, Karras J, Pow-Sang J, Jove R. Mora LB, et al. Cancer Res. 2002 Nov 15;62(22):6659-66. Cancer Res. 2002. PMID: 12438264 - STAT proteins as novel targets for cancer drug discovery.
Turkson J. Turkson J. Expert Opin Ther Targets. 2004 Oct;8(5):409-22. doi: 10.1517/14728222.8.5.409. Expert Opin Ther Targets. 2004. PMID: 15469392 Review. - STAT proteins: novel molecular targets for cancer drug discovery.
Turkson J, Jove R. Turkson J, et al. Oncogene. 2000 Dec 27;19(56):6613-26. doi: 10.1038/sj.onc.1204086. Oncogene. 2000. PMID: 11426647 Review.
Cited by
- Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts.
Zhang X, Yue P, Page BD, Li T, Zhao W, Namanja AT, Paladino D, Zhao J, Chen Y, Gunning PT, Turkson J. Zhang X, et al. Proc Natl Acad Sci U S A. 2012 Jun 12;109(24):9623-8. doi: 10.1073/pnas.1121606109. Epub 2012 May 23. Proc Natl Acad Sci U S A. 2012. PMID: 22623533 Free PMC article. - Therapeutic modulators of STAT signalling for human diseases.
Miklossy G, Hilliard TS, Turkson J. Miklossy G, et al. Nat Rev Drug Discov. 2013 Aug;12(8):611-29. doi: 10.1038/nrd4088. Nat Rev Drug Discov. 2013. PMID: 23903221 Free PMC article. Review. - Stat3 inhibitor Stattic exhibits potent antitumor activity and induces chemo- and radio-sensitivity in nasopharyngeal carcinoma.
Pan Y, Zhou F, Zhang R, Claret FX. Pan Y, et al. PLoS One. 2013;8(1):e54565. doi: 10.1371/journal.pone.0054565. Epub 2013 Jan 29. PLoS One. 2013. PMID: 23382914 Free PMC article. - Signal transducer and activator of transcription 3 is involved in cell growth and survival of human rhabdomyosarcoma and osteosarcoma cells.
Chen CL, Loy A, Cen L, Chan C, Hsieh FC, Cheng G, Wu B, Qualman SJ, Kunisada K, Yamauchi-Takihara K, Lin J. Chen CL, et al. BMC Cancer. 2007 Jun 28;7:111. doi: 10.1186/1471-2407-7-111. BMC Cancer. 2007. PMID: 17598902 Free PMC article. - A novel small molecule, LLL12, inhibits STAT3 phosphorylation and activities and exhibits potent growth-suppressive activity in human cancer cells.
Lin L, Hutzen B, Li PK, Ball S, Zuo M, DeAngelis S, Foust E, Sobo M, Friedman L, Bhasin D, Cen L, Li C, Lin J. Lin L, et al. Neoplasia. 2010 Jan;12(1):39-50. doi: 10.1593/neo.91196. Neoplasia. 2010. PMID: 20072652 Free PMC article.
References
- Darnell, J. J., Kerr, I. & Stark, G. (1994) Science 264, 1415-1421. - PubMed
- Sartor, C., Dziubinski, M., Yu, C., Jove, R. & Ethier, S. (1997) Cancer Res. 57, 978-987. - PubMed
- Garcia, R., Bowman, T., Niu, G., Yu, H., Minton, S., Muro-Cacho, C., Cox, C., Falcone, R., Fairclough, R., Parsons, S., et al. (2001) Oncogene 20, 2499-2513. - PubMed
- Zhong, Z., Wen, Z. & Darnell, J. (1994) Science 264, 95-98. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous