Accelerated probabilistic inference of RNA structure evolution - PubMed (original) (raw)
Accelerated probabilistic inference of RNA structure evolution
Ian Holmes. BMC Bioinformatics. 2005.
Abstract
Background: Pairwise stochastic context-free grammars (Pair SCFGs) are powerful tools for evolutionary analysis of RNA, including simultaneous RNA sequence alignment and secondary structure prediction, but the associated algorithms are intensive in both CPU and memory usage. The same problem is faced by other RNA alignment-and-folding algorithms based on Sankoff's 1985 algorithm. It is therefore desirable to constrain such algorithms, by pre-processing the sequences and using this first pass to limit the range of structures and/or alignments that can be considered.
Results: We demonstrate how flexible classes of constraint can be imposed, greatly reducing the computational costs while maintaining a high quality of structural homology prediction. Any score-attributed context-free grammar (e.g. energy-based scoring schemes, or conditionally normalized Pair SCFGs) is amenable to this treatment. It is now possible to combine independent structural and alignment constraints of unprecedented general flexibility in Pair SCFG alignment algorithms. We outline several applications to the bioinformatics of RNA sequence and structure, including Waterman-Eggert N-best alignments and progressive multiple alignment. We evaluate the performance of the algorithm on test examples from the RFAM database.
Conclusion: A program, Stemloc, that implements these algorithms for efficient RNA sequence alignment and structure prediction is available under the GNU General Public License.
Figures
Figure 1
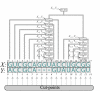
A parse tree for the grammar of Table 1. Each internal node is labeled with a nonterminal (Stem or Loop); additionally, the subsequences (X ij, Y kl) generated by each internal node are shown. The parse tree determines both the structure and alignment of the two sequences. The cut-points of the alignment are the sequence co-ordinates at which the alignment can be split, i.e. {(0, 0), (1, 1), (2, 2) ... (15, 12), (16, 13), (17, 14)}.
Figure 2
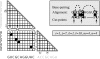
Parsing a pair of sequences (X, Y) using the Inside algorithm involves iterating over subsequence-pairs (X ij, Y kl) specified by four indices (i, j, k, l). In the constrained Inside algorithm, these indices are only valid if the fold envelopes (triangular grids) include the respective subsequences (i, j) and (k, l) (shown as black circles) and the alignment envelope (rectangular grid) includes both cutpoints (i, k) and (j, l) (shown as short diagonal lines). The filled cells in the rectangular grid show the aligned nucleotides. Note that the co-ordinates (i, j, k, l) lie on the grid-lines between the nucleotides.
Figure 3
Bifurcation rules allow a subsequence-pair (X ij, Y kl) to be composed from two adjoining subsequence-pairs (X im, Y kn) and (X nj, Y ni). For this to be permitted by the constraints, the _X_-fold envelope (upper triangular grid) must contain subsequences (i, m), (m, j) and (i, j) (black dots), the _Y_-fold envelope (rightmost triangular grid) must contain subsequences (k, n), (n, l) and (k, l) (black dots) and the alignment envelope (rectangular grid) must contain cutpoints (i, k), (m, n) and (j, l) (short diagonal lines). The filled cells in the rectangular grid show the nucleotide homologies highlighted in the alignment. Note that all co-ordinates (i, j, k, l, m, n) lie on the grid-lines between nucleotides.
Figure 4
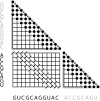
These fold envelopes (triangular grids) limit the maximum length of subsequences (black dots), while the alignment envelope (rectangular grid) limits the maximum deviation of cutpoints (short diagonal lines) from the main diagonal.
Figure 5
These fold envelopes (triangular grids) and alignment envelope (rectangular grid) limit the subsequences (black dots) and cutpoints (short diagonal lines) to those consistent with a given alignment and consensus secondary structure (shown). The alignment path is also shown on the alignment envelope as a solid black line, broken by cutpoints.
Figure 6
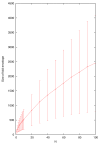
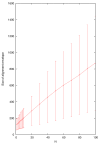
Fold envelope size is highly correlated with N in the _N_-best fold test, although the variance is large due to the diversity of alignments in the test.
Figure 7
Alignment envelope size is highly correlated with N in the _N_-best alignment test, although the variance is large due to the diversity of alignments in the test.
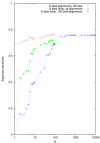
Figure 8
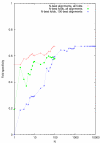
Alignment sensitivity as a function of envelope size parameter N for three different test regimes.
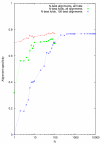
Figure 9
Alignment specificity as a function of envelope size parameter N for three different test regimes.
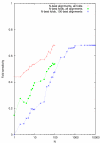
Figure 10
Fold sensitivity as a function of envelope size parameter N for three different test regimes.
Figure 11
Fold specificity as a function of envelope size parameter N for three different test regimes.
Figure 12
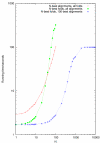
Total running time of stemloc (including envelope generation phases) as a function of envelope size parameter N for three different test regimes.
Figure 13
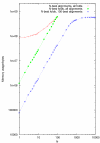
Peak memory usage of stemloc (i.e. the size of the principal CYK matrix) as a function of envelope size parameter N for three different test regimes.
Similar articles
- Pairwise RNA structure comparison with stochastic context-free grammars.
Holmes I, Rubin GM. Holmes I, et al. Pac Symp Biocomput. 2002:163-74. doi: 10.1142/9789812799623_0016. Pac Symp Biocomput. 2002. PMID: 11928472 - CONTRAfold: RNA secondary structure prediction without physics-based models.
Do CB, Woods DA, Batzoglou S. Do CB, et al. Bioinformatics. 2006 Jul 15;22(14):e90-8. doi: 10.1093/bioinformatics/btl246. Bioinformatics. 2006. PMID: 16873527 - Efficient pairwise RNA structure prediction and alignment using sequence alignment constraints.
Dowell RD, Eddy SR. Dowell RD, et al. BMC Bioinformatics. 2006 Sep 4;7:400. doi: 10.1186/1471-2105-7-400. BMC Bioinformatics. 2006. PMID: 16952317 Free PMC article. - Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.
Washietl S, Bernhart SH, Kellis M. Washietl S, et al. Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review. - The art of editing RNA structural alignments.
Andersen ES. Andersen ES. Methods Mol Biol. 2014;1097:379-94. doi: 10.1007/978-1-62703-709-9_17. Methods Mol Biol. 2014. PMID: 24639168 Review.
Cited by
- RNAspa: a shortest path approach for comparative prediction of the secondary structure of ncRNA molecules.
Horesh Y, Doniger T, Michaeli S, Unger R. Horesh Y, et al. BMC Bioinformatics. 2007 Oct 1;8:366. doi: 10.1186/1471-2105-8-366. BMC Bioinformatics. 2007. PMID: 17908318 Free PMC article. - SPARSE: quadratic time simultaneous alignment and folding of RNAs without sequence-based heuristics.
Will S, Otto C, Miladi M, Möhl M, Backofen R. Will S, et al. Bioinformatics. 2015 Aug 1;31(15):2489-96. doi: 10.1093/bioinformatics/btv185. Epub 2015 Apr 2. Bioinformatics. 2015. PMID: 25838465 Free PMC article. - Evolutionary triplet models of structured RNA.
Bradley RK, Holmes I. Bradley RK, et al. PLoS Comput Biol. 2009 Aug;5(8):e1000483. doi: 10.1371/journal.pcbi.1000483. Epub 2009 Aug 28. PLoS Comput Biol. 2009. PMID: 19714212 Free PMC article. - Comparative genomics beyond sequence-based alignments: RNA structures in the ENCODE regions.
Torarinsson E, Yao Z, Wiklund ED, Bramsen JB, Hansen C, Kjems J, Tommerup N, Ruzzo WL, Gorodkin J. Torarinsson E, et al. Genome Res. 2008 Feb;18(2):242-51. doi: 10.1101/gr.6887408. Epub 2007 Dec 20. Genome Res. 2008. PMID: 18096747 Free PMC article. - An efficient genetic algorithm for structural RNA pairwise alignment and its application to non-coding RNA discovery in yeast.
Taneda A. Taneda A. BMC Bioinformatics. 2008 Dec 5;9:521. doi: 10.1186/1471-2105-9-521. BMC Bioinformatics. 2008. PMID: 19061486 Free PMC article.