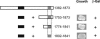Sites of proteolytic processing and noncovalent association of the distal C-terminal domain of CaV1.1 channels in skeletal muscle - PubMed (original) (raw)
Sites of proteolytic processing and noncovalent association of the distal C-terminal domain of CaV1.1 channels in skeletal muscle
Joanne T Hulme et al. Proc Natl Acad Sci U S A. 2005.
Abstract
In skeletal muscle cells, voltage-dependent potentiation of Ca2+ channel activity requires phosphorylation by cAMP-dependent protein kinase (PKA) anchored via an A-kinase anchoring protein (AKAP15), and the most rapid sites of phosphorylation are located in the C-terminal domain. Surprisingly, the site of interaction of the complex of PKA and AKAP15 with the alpha1-subunit of Ca(V)1.1 channels lies in the distal C terminus, which is cleaved from the remainder of the channel by in vivo proteolytic processing. Here we report that the distal C terminus is noncovalently associated with the remainder of the channel via an interaction with a site in the proximal C-terminal domain when expressed as a separate protein in mammalian nonmuscle cells. Deletion mapping of the C terminus of the alpha1-subunit using the yeast two-hybrid assay revealed that a distal C-terminal peptide containing amino acids 1802-1841 specifically interacts with a region in the proximal C terminus containing amino acid residues 1556-1612. Analysis of the purified alpha1-subunit of Ca(V)1.1 channels from skeletal muscle by saturation sequencing of the intracellular peptides by tandem mass spectrometry identified the site of proteolytic processing as alanine 1664. Our results support the conclusion that a noncovalently associated complex of the alpha1-subunit truncated at A1664 with the proteolytically cleaved distal C-terminal domain, AKAP15, and PKA is the primary physiological form of Ca(V)1.1 channels in skeletal muscle cells.
Figures
Fig. 1.
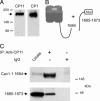
Coimmunoprecipitation of the separately expressed distal C terminus with truncated CaV1.1 channels in tsA-201 cells. (A) Immunoblot showing that expression of CaV1.1 channels by transfection of full-length cDNA in tsA-201 cells yields only full-length α1-subunit proteins, as detected by an anti-peptide antibody against the final C-terminal peptide (anti-CP1). (B) Schematic diagram of truncated CaV1.1 channels and the separately expressed distal C terminus used in coimmunoprecipitation experiments. (C) Lysates of tsA-201 cells transfected with CaV1.1 1684 and the distal C terminus (1685–1873) were immunoprecipitated with anti-CP11 (lane 2) or control IgG (lane 3). Immunoblots were probed with anti-CP11 (Upper) or anti-myc (Lower). Positive control for immunoblotting was 20 μl of lysate (lane 1).
Fig. 2.
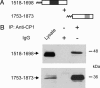
Coimmunoprecipitation of the distal and proximal C-terminal domains of CaV1.1 channels expressed in tsA-201 cells. (A) Schematic map of the lipid-anchored CaV1.1 C-terminal constructs examined in coimmunoprecipitation experiments. (B) TsA-201 cells were cotransfected with the constructs depicted in A, and cell lysates were immunoprecipitated with anti-CP1 or control IgG. Immunoblots were probed with anti-CP11 (Upper) or anti-CP1 (Lower) to detect immunoprecipitated CaV1.1 1518–1698 and CaV1.1 1753–1873 protein, respectively.
Fig. 3.
Amino acid residues 1556–1612 in the proximal C terminus of CaV1.1 bind the distal C terminus. (Left) Schematic map of the proximal C-terminal constructs of CaV1.1 that were cloned into the pAS2.1 vector and cotransformed with the distal C terminus (1753–1873 pACT2) or pACT2 alone into the Y190 yeast strain. Numbers correspond to the amino acid residues in CaV1.1. (Right) Representative growth of yeast cotransformed with the CaV1.1 proximal and distal C-terminal plasmids depicted on the left and the resulting β-galactosidase assay (+ or –).
Fig. 4.
Amino acid residues 1802–1841 in the distal C terminus of CaV1.1 bind the proximal C terminus. (Left) Schematic map of the distal C-terminal constructs of CaV1.1 that were cloned into the pACT2 vector and cotransformed with the proximal C terminus (1556–1612 pAS2.1) or pAS2.1 alone into the Y190 yeast strain. Numbers correspond to the amino acid residues in CaV1.1. (Right) Representative growth of yeast cotransformed with the CaV1.1 proximal and distal C-terminal plasmids depicted on the left and the resulting β-galactosidase assay (+ or –).
Fig. 5.
Identification of the C terminus of proteolytically processed CaV1.1 by mass spectrometry. (A) Coomassie blue staining of the CaV1.1 complex. The α1-subunit band was excised from the gel and subjected to “in-gel” proteolysis with three different proteases. The other bands on the gel correspond to the α2-, β-, and γ-subunits that copurify in the CaV1.1 channel complex. (B) Schematic diagram showing the coverage of proteolytic peptides of the α1-subunit of CaV1.1 channels identified by LC/MS/MS. (C) Deduced amino acid sequence of the cleaved CaV1.1 C terminus. The location of the proteolytic peptides obtained from digests with trypsin (black bar), endoproteinase Asp-N (striped bar) and endoproteinase Glu-C (gray bar) are shown.
Fig. 6.
Conserved amino acid sequences of CaV1.1 and CaV1.2 channels at the sites of C-terminal interaction and proteolytic cleavage. Amino acid sequence alignment of the PCID, DCID, PEST motif, the proteolytic cleavage site (asterisk and scissors), and the AKAP15-binding domain (ABD; critical residues shown in bold) identified in CaV1.1 with CaV1.2, CaV1.3, and CaV1.4. Conserved amino acids are shaded in black, and similar amino acids are shaded in gray.
Similar articles
- Autoinhibitory control of the CaV1.2 channel by its proteolytically processed distal C-terminal domain.
Hulme JT, Yarov-Yarovoy V, Lin TW, Scheuer T, Catterall WA. Hulme JT, et al. J Physiol. 2006 Oct 1;576(Pt 1):87-102. doi: 10.1113/jphysiol.2006.111799. Epub 2006 Jun 29. J Physiol. 2006. PMID: 16809371 Free PMC article. - Beta-adrenergic regulation requires direct anchoring of PKA to cardiac CaV1.2 channels via a leucine zipper interaction with A kinase-anchoring protein 15.
Hulme JT, Lin TW, Westenbroek RE, Scheuer T, Catterall WA. Hulme JT, et al. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):13093-8. doi: 10.1073/pnas.2135335100. Epub 2003 Oct 20. Proc Natl Acad Sci U S A. 2003. PMID: 14569017 Free PMC article. - A novel leucine zipper targets AKAP15 and cyclic AMP-dependent protein kinase to the C terminus of the skeletal muscle Ca2+ channel and modulates its function.
Hulme JT, Ahn M, Hauschka SD, Scheuer T, Catterall WA. Hulme JT, et al. J Biol Chem. 2002 Feb 8;277(6):4079-87. doi: 10.1074/jbc.M109814200. Epub 2001 Nov 30. J Biol Chem. 2002. PMID: 11733497 - Regulation of Cardiac Calcium Channels in the Fight-or-Flight Response.
Catterall WA. Catterall WA. Curr Mol Pharmacol. 2015;8(1):12-21. doi: 10.2174/1874467208666150507103417. Curr Mol Pharmacol. 2015. PMID: 25966697 Free PMC article. Review. - Targeting mechanisms of high voltage-activated Ca2+ channels.
Herlitze S, Xie M, Han J, Hümmer A, Melnik-Martinez KV, Moreno RL, Mark MD. Herlitze S, et al. J Bioenerg Biomembr. 2003 Dec;35(6):621-37. doi: 10.1023/b:jobb.0000008027.19384.c0. J Bioenerg Biomembr. 2003. PMID: 15000523 Review.
Cited by
- Signaling complexes of voltage-gated sodium and calcium channels.
Catterall WA. Catterall WA. Neurosci Lett. 2010 Dec 10;486(2):107-16. doi: 10.1016/j.neulet.2010.08.085. Epub 2010 Sep 17. Neurosci Lett. 2010. PMID: 20816922 Free PMC article. Review. - Sequence differences in the IQ motifs of CaV1.1 and CaV1.2 strongly impact calmodulin binding and calcium-dependent inactivation.
Ohrtman J, Ritter B, Polster A, Beam KG, Papadopoulos S. Ohrtman J, et al. J Biol Chem. 2008 Oct 24;283(43):29301-11. doi: 10.1074/jbc.M805152200. Epub 2008 Aug 21. J Biol Chem. 2008. PMID: 18718913 Free PMC article. - Molecular determinants of the CaVbeta-induced plasma membrane targeting of the CaV1.2 channel.
Bourdin B, Marger F, Wall-Lacelle S, Schneider T, Klein H, Sauvé R, Parent L. Bourdin B, et al. J Biol Chem. 2010 Jul 23;285(30):22853-63. doi: 10.1074/jbc.M110.111062. Epub 2010 May 17. J Biol Chem. 2010. PMID: 20478999 Free PMC article. - Deletion of the distal C terminus of CaV1.2 channels leads to loss of beta-adrenergic regulation and heart failure in vivo.
Fu Y, Westenbroek RE, Yu FH, Clark JP 3rd, Marshall MR, Scheuer T, Catterall WA. Fu Y, et al. J Biol Chem. 2011 Apr 8;286(14):12617-26. doi: 10.1074/jbc.M110.175307. Epub 2011 Jan 7. J Biol Chem. 2011. PMID: 21216955 Free PMC article. - Ca(V)1.1: The atypical prototypical voltage-gated Ca²⁺ channel.
Bannister RA, Beam KG. Bannister RA, et al. Biochim Biophys Acta. 2013 Jul;1828(7):1587-97. doi: 10.1016/j.bbamem.2012.09.007. Epub 2012 Sep 13. Biochim Biophys Acta. 2013. PMID: 22982493 Free PMC article. Review.
References
- Catterall, W. A. (1991) Cell 64, 871–874. - PubMed
- Adams, B. A. & Beam, K. G. (1990) FASEB J. 4, 2809–2816. - PubMed
- Rios, E. & Pizarro, G. (1991) Physiol. Rev. 71, 849–908. - PubMed
- Sculptoreanu, A., Scheuer, T. & Catterall, W. A. (1993) Nature 364, 240–243. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous