Inhibition of Epstein-Barr virus OriP function by tankyrase, a telomere-associated poly-ADP ribose polymerase that binds and modifies EBNA1 - PubMed (original) (raw)
Inhibition of Epstein-Barr virus OriP function by tankyrase, a telomere-associated poly-ADP ribose polymerase that binds and modifies EBNA1
Zhong Deng et al. J Virol. 2005 Apr.
Abstract
Tankyrase (TNKS) is a telomere-associated poly-ADP ribose polymerase (PARP) that has been implicated along with several telomere repeat binding factors in the regulation of Epstein-Barr virus origin of plasmid replication (OriP). We now show that TNKS1 can bind to the family of repeats (FR) and dyad symmetry regions of OriP by using a chromatin immunoprecipitation assay and DNA affinity purification. TNKS1 and TNKS2 bound to EBNA1 in coimmunoprecipitation experiments with transfected cell lysates and with purified recombinant proteins in vitro. Two RXXPDG-like TNKS-interacting motifs in the EBNA1 amino-terminal domain mediated binding with the ankyrin repeat domain of TNKS. Mutations of both motifs at EBNA1 G81 and G425 abrogated TNKS binding and enhanced EBNA1-dependent replication of OriP. Small hairpin RNA targeted knock-down of TNKS1 enhanced OriP-dependent DNA replication. Overexpression of TNKS1 or TNKS2 inhibited OriP-dependent DNA replication, while a PARP-inactive form of TNKS2 (M1045V) was compromised for this inhibition. We show that EBNA1 is subject to PAR modification in vivo and to TNKS1-mediated PAR modification in vitro. These results indicate that TNKS proteins can interact directly with the EBNA1 protein, associate with the FR region of OriP in vivo, and inhibit OriP replication in a PARP-dependent manner.
Figures
FIG. 1.
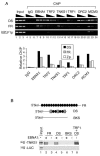
TNKS binds the FR region of OriP. (A) Raji cells were subjected to ChIP assay with control IgG (lanes 4 to 6) or specific antibodies against EBNA1 (lanes 7 to 9), TRF2 (lanes 10 to 12), TNKS1 (lanes 13 to 15), TRF1 (lanes 16 to 18), ORC2 (lanes 19 to 21), or MCM3 (lane 22 to 24). ChIP DNA was amplified for EBV regions DS, FR, and BZLF1p as indicated. Input (lanes 1 to 3) and chromatin immunoprecipitated DNA were serially diluted threefold (lanes 1 to 3). Quantification of the average intensity of ethidium bromide-stained DNA products was done relative to total input DNA and shown in the bar graph below. (B) FR, DS, or BKS DNA affinity binding was assayed by using in vitro-translated 35S-TNKS1 or 35S-luciferase (35S-LUC) control protein. Binding was measured with (+) or without (−) recombinant baculovirus-expressed and purified EBNA1. For the positive control, recombinant TRF1 was used instead of EBNA1 (lane 8). The schematic above depicts biotinylated (Bio) templates with EBNA1 sites (circles) and TRF binding sites (squares).
FIG. 2.
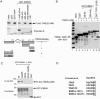
TNKS binds EBNA1 directly. (A) 293 cells were cotransfected with plasmids expressing EBNA1 and either FLAG-TNKS1 (top panel), FLAG-TNKS2 (middle panel), or FLAG-TNKS2-ANK (lower panel). Immunoprecipitates using rabbit anti-EBNA1, anti-FLAG, or control rabbit IgG were assayed by Western blotting with monoclonal antibodies specific for FLAG or EBNA1. (B) Cell extracts containing full-length FLAG-TNKS1 (top panel) or FLAG-TNKS2 (lower panel) were assayed for binding GST or GST-EBNA1(1-440ΔGA). Bound proteins were assayed by Western blotting with the anti-TNKS antibodies T1S (upper panel) and T12 (lower panel). (C) Purified baculovirus-expressed TNKS1 and EBNA1 form a complex in vitro. EBNA1 alone (lanes 3, 6, and 9), TNKS alone (lanes 4, 7, and 10), or the two proteins mixed in vitro (lanes 5, 8, and 11) were subjected to immunoprecipitation (IP) with IgG (lanes 3 to 5) or antibodies against EBNA1 (lanes 6 to 8) or TNKS1 (lanes 9 to 11). Immunoprecipitates were washed extensively and assayed by Western immunoblotting (IB) with monoclonal antibodies to TNKS1 (top panel) or EBNA1 (bottom panel). Inputs (lanes 1 and 2) represent 5% of the total material used for immunoprecipitates.
FIG. 3.
TNKS binds to two RXXPDG-like motifs in EBNA1. (A) Extracts from 293 cells transfected with the FLAG-tagged ankyrin domain of TNKS2 (FLAG-TNKS2-ANK) were used to assay binding to GST fusions of various EBNA1 subdomains, as indicated above. Bound proteins were detected by anti-FLAG immunoblotting (top panel). GST proteins were visualized by Ponceaus S staining of nitrocellulose blots (lower panel). GST-IRAP is a positive control for TNKS2 binding. A summary of several independent binding experiments is given in the schematic below. (B) Purified GST (lanes 3 and 4), GST-EBNA1(58-93) (lanes 5 and 6), or GST-EBNA1(339-449) (lanes 7 and 8) were incubated with (+) or without (−) the purified TNKS1 ankyrin repeat domain (aa 641 to 831). Glutathione agarose-bound proteins were visualized by Coomassie blue staining in SDS-PAGE. (C) GST-EBNA1(1-440ΔGA) containing substitution mutations at G425, G81, or both was assayed for binding to TNKS2 as described for panel A. Coomassie staining of GST-EBNA1 proteins is shown below. (D) Alignment of the RXXPDG-like motifs from EBNA1 and other TNKS-interacting proteins.
FIG. 4.
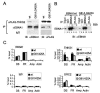
EBNA1 (G81A/G425A) diminishes TNKS binding in vivo. (A) wt or G81A/G425A EBNA1 was coexpressed with FLAG-TNKS2 in HeLa cells and assayed for coimmunoprecipitation. Immunoprecipitation with anti-FLAG, anti-EBNA1, or IgG control was analyzed by immunoblotting with anti-EBNA1 (left panel) or anti-FLAG (right panel). (B) wt or G81A/G425A EBNA1 was coexpressed with FLAG-TNKS2 in HeLa cells and assayed for coimmunoprecipitation in the absence (−) or presence (+) of 100 μg of EtBr/ml. Immunoprecipitates were analyzed by immunoblotting with anti-EBNA1. (C) wt (black fill) or G81A/G425A (grey fill) EBNA1 was coexpressed in HeLa cells with OriP-containing plasmid and assayed by ChIP for amplification of DS, FR, ampicillin gene (Amp), or cellular actin DNA, using antibody to EBNA1, TNKS1, ORC2, or IgG.
FIG. 5.
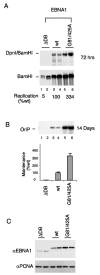
Mutation of the EBNA1 TNKS interaction motifs enhances replication and plasmid maintenance. (A) EBNA1-dependent DNA replication was assayed in HeLa cells transfected with OriP plasmids expressing EBNA1 lacking its DNA binding domain (ΔDB), EBNA1 (wt), or EBNA1 (G81A/G425A), as indicated above each lane. Dpn1 and BamHI double cuts (top panel) indicate replicated DNA relative to the total recovered DNA linearized with BamHI alone (bottom panel). Replication activity was quantified for at least three independent experiments, and standard deviations were less than 15%. (B) Plasmid maintenance was assayed for OriP plasmids encoding ΔDB (lanes 1 and 2), wt (lanes 3 and 4), or G81/425A (lanes 5 and 6) EBNA1. Recovered plasmid was linearized with BamHI and detected by Southern hybridization. Quantification of three independent experiments is presented in the bar graph below. (C) Western blot analysis of hemagglutinin-tagged EBNA1 derivatives from transfected cell extracts described for panel A above.
FIG. 6.
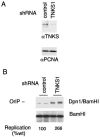
shRNA-mediated depletion of TNKS1 enhances OriP replication. EBNA1-positive ZKO-293 cells were cotransfected with OriP plasmid and either shRNA control plasmid or shRNA target plasmid for TNKS1. (A) Seventy-two hours posttransfection, cell lysates were assayed by Western blotting for expression of TNKS1 (top panel) or control PCNA protein (bottom panel). (B) The same transfected cells were assayed for OriP-dependent replication of transfected plasmids. Quantification of replication was the average from three independent experiments.
FIG. 7.
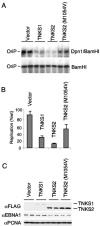
Ectopic expression of TNKS proteins inhibits OriP replication. (A) EBNA1-positive ZKO-293 cells were cotransfected with OriP plasmid and expression vector for TNKS1, TNKS2, or a PARP-defective mutant of TNKS2 (M1054V), as indicated above each lane. The transfected cells were assayed for OriP-dependent DNA replication. (B) Quantification of OriP DNA replication represents the average for three independent experiments. (C) Western blot analysis of transfected cell extracts shown above. Antibodies to FLAG-tagged TNKS1 or TNKS2 (top panel), EBNA1 (middle panel), and control protein PCNA (bottom panel) are indicated.
FIG. 8.
PAR modification of EBNA1. (A) EBV-positive Raji cells were immunoprecipitated (IP) with anti-EBNA1, anti-PAR, or IgG control and then analyzed by immunoblotting (IB) with anti-EBNA1 (left panel), anti-PAR (middle panel), or anti-TNKS1 (right panel). Immunoprecipitated proteins were eluted at 65°C in Laemmli buffer. The positions of EBNA1 and TNKS1 proteins are indicated by the arrows. The asterisk indicates a cross-reacting band of unknown identity. (B) EBV-positive Raji cells left untreated (top panels) or treated with 3-AB for 4 h (lower panels) were subjected to immunoprecipitation with antibodies against EBNA1, PAR, or control mouse IgG. Immunoprecipitates were immunoblotted with anti-EBNA1 antibodies (left panels) or anti-PAR antibodies (right panels). Immunoprecipitated proteins were eluted with Laemmli buffer at 95°C for 5 min. (C) Purified recombinant TNKS1 protein was incubated with purified recombinant EBNA1 with (+) or without (−) 1 mM NAD+ as indicated and assayed by Western blotting with anti-EBNA1 (top) or anti-PAR (lower panel) antibodies.
Similar articles
- Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication.
Deng Z, Lezina L, Chen CJ, Shtivelband S, So W, Lieberman PM. Deng Z, et al. Mol Cell. 2002 Mar;9(3):493-503. doi: 10.1016/s1097-2765(02)00476-8. Mol Cell. 2002. PMID: 11931758 - Regulation of Epstein-Barr virus OriP replication by poly(ADP-ribose) polymerase 1.
Tempera I, Deng Z, Atanasiu C, Chen CJ, D'Erme M, Lieberman PM. Tempera I, et al. J Virol. 2010 May;84(10):4988-97. doi: 10.1128/JVI.02333-09. Epub 2010 Mar 10. J Virol. 2010. PMID: 20219917 Free PMC article. - Structural Basis for Cooperative Binding of EBNA1 to the Epstein-Barr Virus Dyad Symmetry Minimal Origin of Replication.
Malecka KA, Dheekollu J, Deakyne JS, Wiedmer A, Ramirez UD, Lieberman PM, Messick TE. Malecka KA, et al. J Virol. 2019 Sep 30;93(20):e00487-19. doi: 10.1128/JVI.00487-19. Print 2019 Oct 15. J Virol. 2019. PMID: 31142669 Free PMC article. - Replication licensing of the EBV oriP minichromosome.
Hirai K, Shirakata M. Hirai K, et al. Curr Top Microbiol Immunol. 2001;258:13-33. doi: 10.1007/978-3-642-56515-1_2. Curr Top Microbiol Immunol. 2001. PMID: 11443858 Review. - EBNA1 and host factors in Epstein-Barr virus latent DNA replication.
Frappier L. Frappier L. Curr Opin Virol. 2012 Dec;2(6):733-9. doi: 10.1016/j.coviro.2012.09.005. Epub 2012 Sep 30. Curr Opin Virol. 2012. PMID: 23031715 Review.
Cited by
- At a crossroads: human DNA tumor viruses and the host DNA damage response.
Nikitin PA, Luftig MA. Nikitin PA, et al. Future Virol. 2011 Jul;6(7):813-830. doi: 10.2217/fvl.11.55. Future Virol. 2011. PMID: 21927617 Free PMC article. - NAD+ Degrading Enzymes, Evidence for Roles During Infection.
Tan A, Doig CL. Tan A, et al. Front Mol Biosci. 2021 Aug 16;8:697359. doi: 10.3389/fmolb.2021.697359. eCollection 2021. Front Mol Biosci. 2021. PMID: 34485381 Free PMC article. Review. - Telomeres and viruses: common themes of genome maintenance.
Deng Z, Wang Z, Lieberman PM. Deng Z, et al. Front Oncol. 2012 Dec 31;2:201. doi: 10.3389/fonc.2012.00201. eCollection 2012. Front Oncol. 2012. PMID: 23293769 Free PMC article. - The EBNA1 protein of Epstein-Barr virus functionally interacts with Brd4.
Lin A, Wang S, Nguyen T, Shire K, Frappier L. Lin A, et al. J Virol. 2008 Dec;82(24):12009-19. doi: 10.1128/JVI.01680-08. Epub 2008 Oct 15. J Virol. 2008. PMID: 18922874 Free PMC article. - Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes.
Gagné JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Gagné JP, et al. Nucleic Acids Res. 2008 Dec;36(22):6959-76. doi: 10.1093/nar/gkn771. Epub 2008 Nov 3. Nucleic Acids Res. 2008. PMID: 18981049 Free PMC article.
References
- Ame, J. C., C. Spenlehauer, and G. De Murcia. 2004. The PARP superfamily. Bioessays 26:882-893. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources