The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4 - PubMed (original) (raw)
The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4
Michael K Baxter et al. J Virol. 2005 Apr.
Abstract
The papillomavirus transcriptional activator, E2, is involved in key functions of the viral life cycle. These include transcriptional regulation, viral DNA replication, and viral genome segregation. The transactivation domain of E2 is required for each of these functions. To identify the regions of the domain that mediate binding to mitotic chromosomes, a panel of mutations has been generated and their effect on various E2 functions has been analyzed. A structural model of the bovine papillomavirus type 1 (BPV1) E2 transactivation domain was generated based on its homology with the solved structure of the human papillomavirus type 16 (HPV16) domain. This model was used to identify distinct surfaces of the domain to be targeted by point mutation to further delineate the functional region of the transactivation domain responsible for mitotic chromosome association. The mutated E2 proteins were assessed for mitotic chromosome binding and, in addition, transcriptional activation and transcriptional repression activities. Mutation of amino acids R37 and I73, which are located on a surface of the domain that in HPV16 E2 is reported to mediate self-interaction, completely eliminated mitotic chromosome binding. Mitotic chromosome binding activity was found to correlate well with the ability to interact with the cellular chromosomal associated factor Brd4, which has recently been proposed to mediate the association between BPV1 E2 and mitotic chromosomes.
Figures
FIG. 1.
Structural model of BPV1 E2 transactivation domain. Shown are opposing views of a structural representation of the BPV1 E2 transactivation domain. The BPV transactivation domain (amino acids 1 to 203) was modeled on the structure of the HPV16 transactivation domain by using the programs Look and RasMol. The model is displayed in space-filling mode, with the eight mutation target groups (A to I) indicated in different colors. The locations of the amino (N) and carboxyl (C) termini of the transactivation domain are indicated in both views of the structural model.
FIG. 2.
Expression of mutated E2 proteins. Shown is a Western blot of protein extracts from CV-1 cell lines stably transfected with pMEP4 (No E2), wild-type pMEP-E2 (WT E2), or mutated pMEP-E2 plasmids (A1 to I2) (specific mutations are listed in Table 1). E2 proteins were detected with the anti-E2 monoclonal antibody B201.
FIG.3.
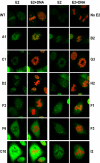
Mitotic chromosome binding of mutated E2 proteins. E2 localization was analyzed in stably transfected CV-1 cell lines by confocal fluorescence microscopy. Shown are representative mitotic cells for the indicated samples (mutations are listed in Table 1). The sample without E2 (No E2) was transfected with the pMEP4 vector. E2 was detected by indirect immunofluorescence with an FITC-conjugated secondary antibody and is shown in green (E2), while the merged image shows E2 staining plus cellular DNA counterstained in red with propidium iodide (E2+DNA). The images are ordered from those that bind chromosomes at the top to those that don't at the bottom. WT, wild type
FIG. 4.
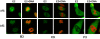
The chromosome binding phenotype of several E2 proteins is temperature sensitive. E2 localization was analyzed in stably transfected CV-1 cell lines that were cultured at 34 or 37°C by confocal fluorescence microscopy. Shown are representative mitotic cells for the B3, C9, and F3 proteins. E2 was detected by indirect immunofluorescence with an FITC-conjugated secondary antibody and is shown in green (E2), while the merged image shows E2 staining plus cellular DNA counterstained in red with propidium iodide (E2+DNA).
FIG. 5.
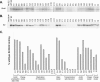
Brd4 interaction of mutated E2 proteins. (A) In vitro-translated E2 proteins were mixed with 35S-labeled in vitro-translated Brd4 and immunoprecipitated with the E2-specific antibody B202. Shown is a representative autoradiograph of the SDS-PAGE analysis of these proteins. The location of the full-length Brd4 band is indicated on the left. (B) 35S-labeled in vitro-translated E2 proteins were mixed with a protein extract containing baculovirus-expressed FLAG-Brd4 and immunoprecipitated with anti-FLAG M2 agarose affinity gel. Shown is an autoradiograph of SDS-PAGE analysis of the precipitated proteins. The location of the E2 band is indicated on the left. (C) Quantitation of Brd4-E2 binding experiments. The average of binding data is derived from experiments as described for panel B. Brd4 binding activity is expressed relative to that of wild-type (WT) E2, which has been set at 100%.
FIG. 6.
HeLa cell growth suppression activity of mutated E2 proteins. HeLa cells were transfected with pMEP4 (No E2), wild-type pMEP-E2 (WT E2), or mutated pMEP-E2 constructs as indicated. Transfected cells were selected with 200 μg of hygromycin B/ml and cultured at either 34 or 37°C, as indicated. When the No E2 plate approached confluence, the cells were fixed and stained with methylene blue. Shown are plates from several groups.
FIG. 7.
Location of key mutations on the transactivation domain. The positions of the R37 and I73 residues, which are key for mitotic chromosome binding, are shown on models of both a monomer and homodimer of the BPV1 E2 transactivation domain.
Similar articles
- The papillomavirus E2 proteins.
McBride AA. McBride AA. Virology. 2013 Oct;445(1-2):57-79. doi: 10.1016/j.virol.2013.06.006. Epub 2013 Jul 10. Virology. 2013. PMID: 23849793 Free PMC article. Review. - Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function.
Schweiger MR, You J, Howley PM. Schweiger MR, et al. J Virol. 2006 May;80(9):4276-85. doi: 10.1128/JVI.80.9.4276-4285.2006. J Virol. 2006. PMID: 16611886 Free PMC article. - Conditional mutations in the mitotic chromosome binding function of the bovine papillomavirus type 1 E2 protein.
Zheng PS, Brokaw J, McBride AA. Zheng PS, et al. J Virol. 2005 Feb;79(3):1500-9. doi: 10.1128/JVI.79.3.1500-1509.2005. J Virol. 2005. PMID: 15650176 Free PMC article. - Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses.
McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. McPhillips MG, et al. J Virol. 2006 Oct;80(19):9530-43. doi: 10.1128/JVI.01105-06. J Virol. 2006. PMID: 16973557 Free PMC article. - Involvement of Brd4 in different steps of the papillomavirus life cycle.
Iftner T, Haedicke-Jarboui J, Wu SY, Chiang CM. Iftner T, et al. Virus Res. 2017 Mar 2;231:76-82. doi: 10.1016/j.virusres.2016.12.006. Epub 2016 Dec 10. Virus Res. 2017. PMID: 27965149 Free PMC article. Review.
Cited by
- Molecular mechanisms of human papillomavirus-related carcinogenesis in head and neck cancer.
Faraji F, Zaidi M, Fakhry C, Gaykalova DA. Faraji F, et al. Microbes Infect. 2017 Sep-Oct;19(9-10):464-475. doi: 10.1016/j.micinf.2017.06.001. Epub 2017 Jun 12. Microbes Infect. 2017. PMID: 28619685 Free PMC article. Review. - Genome-wide Profiling Reveals Remarkable Parallels Between Insertion Site Selection Properties of the MLV Retrovirus and the piggyBac Transposon in Primary Human CD4(+) T Cells.
Gogol-Döring A, Ammar I, Gupta S, Bunse M, Miskey C, Chen W, Uckert W, Schulz TF, Izsvák Z, Ivics Z. Gogol-Döring A, et al. Mol Ther. 2016 Mar;24(3):592-606. doi: 10.1038/mt.2016.11. Epub 2016 Jan 12. Mol Ther. 2016. PMID: 26755332 Free PMC article. - The papillomavirus E2 proteins.
McBride AA. McBride AA. Virology. 2013 Oct;445(1-2):57-79. doi: 10.1016/j.virol.2013.06.006. Epub 2013 Jul 10. Virology. 2013. PMID: 23849793 Free PMC article. Review. - Abrogation of the Brd4-positive transcription elongation factor B complex by papillomavirus E2 protein contributes to viral oncogene repression.
Yan J, Li Q, Lievens S, Tavernier J, You J. Yan J, et al. J Virol. 2010 Jan;84(1):76-87. doi: 10.1128/JVI.01647-09. J Virol. 2010. PMID: 19846528 Free PMC article. - Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function.
Schweiger MR, You J, Howley PM. Schweiger MR, et al. J Virol. 2006 May;80(9):4276-85. doi: 10.1128/JVI.80.9.4276-4285.2006. J Virol. 2006. PMID: 16611886 Free PMC article.
References
- Antson, A. A., J. E. Burns, O. V. Moroz, D. J. Scott, C. M. Sanders, I. B. Bronstein, G. G. Dodson, K. S. Wilson, and N. J. Maitland. 2000. Structure of the intact transactivation domain of the human papillomavirus E2 protein. Nature 403:805-809. - PubMed
- Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 with mitotic chromosomes. Virology 270:124-134. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources