Escape mutations alter proteasome processing of major histocompatibility complex class I-restricted epitopes in persistent hepatitis C virus infection - PubMed (original) (raw)
Escape mutations alter proteasome processing of major histocompatibility complex class I-restricted epitopes in persistent hepatitis C virus infection
Yoichi Kimura et al. J Virol. 2005 Apr.
Abstract
Mutations in hepatitis C virus (HCV) genomes facilitate escape from virus-specific CD8+ T lymphocytes in persistently infected chimpanzees. Our previous studies demonstrated that many of the amino acid substitutions in HCV epitopes prevented T-cell receptor recognition or binding to class I major histocompatibility complex molecules. Here we report that mutations within HCV epitopes also cause their destruction by changing the pattern of proteasome digestion. This mechanism of immune evasion provides further evidence of the potency of CD8+ T-cell selection pressure against HCV and should be considered when evaluating the significance of mutations in viral genomes from persistently infected chimpanzees and humans.
Figures
FIG. 1.
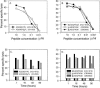
Class I MHC restriction of CD8+ CTL lines from chronically infected animals. CD8+ CTL lines specific for the HCV core (A) or NS4B (B) proteins were derived from the livers of chimpanzees CH-503 and CBO609, respectively. A panel of 721.221 target cells transfected with individual class I Patr molecules from (A) CH-503 or (B) CBO609 were infected overnight with recombinant vaccinia viruses expressing the core (A) or NS4B (B) HCV proteins at an MOI of 10. Cytolysis was measured at the indicated E:T cell ratios in a 4-h assay.
FIG. 2.
Mutations in the core and NS4B proteins of HCV quasispecies from chronically infected animals. HCV RNA was isolated from plasma collected 7 (CH-503) or 5 (CBO609 and CBO603) years after infection with HCV-1/910. cDNA was prepared, and nested PCR primers were used to amplify viral sequences from the core and NS4B genes. PCR products were ligated into plasmids, and multiple molecular clones were sequenced. The C131 and NS4B1723 epitopes are boxed. +ive, positive;-ive, negative.
FIG. 3.
Recognition of target cells sensitized with wild-type or mutant HCV peptides. Patr-B1601 (A)- or A0101 (B)-transfected 721.221 target cells were sensitized for 1 h with various concentrations of nonameric C131 or NS4B1723 wild-type and mutant peptides. They were then cocultured with the (A) C131 or (B) NS4B1723-specific CTL lines at an E:T ratio of 20:1 in a 4-h 51Cr release assay. The target cells were also incubated with the (C) C131 or (D) NS4B1723 peptides (10 μM) for 1 h, washed three times, and then cultured for an additional 1, 16, 48, or 96 h before labeling with 51Cr. Peptide-sensitized targets were then cocultured with the core or NS4-specific CTL lines for 4 h at a 20:1 E:T ratio.
FIG. 4.
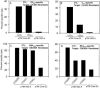
Expression of wild-type and mutated HCV proteins in target cells. Fibroblast cell lines from chimpanzee CH-503 were infected with VVT7 and then transiently transfected with plasmids pTMI(C-E2wt) or pTMI(C-E2/137L) that expressed the wild-type proteins or core proteins with I137L substitution, respectively. They were then incubated with CTL lines specific for the E2588 epitope that was intact in both plasmids (A) or the C131 epitope (B). Fibroblast targets from CBO609 were similarly transfected with plasmids pTMI(NS2-5wt) or pTMI(NS2-5/L1723 or pTMINS2-5/V1723). They were then incubated with a CTL line against the control (i.e., intact) epitope NS4B1939 (C) or the CTL line targeting NS4B1723 (D). Effector and 51Cr-labeled target cells were cocultured at a ratio of 20:1 for 4 h. Results are representative of three replicate experiments.
FIG. 5.
Proteasome processing of synthetic NS4B peptides. (A) Two synthetic peptides spanning amino acids S1712 to R1741 of NS4B containing either the HCV-1 wild-type (M1723) or mutant (L1723) sequences were synthesized and purified by RP-HPLC. NS4B1723 epitopes are boxed. Both peptides were processed with the constitutive proteasomes (CP) or immunoproteasomes (IP) for 16 h and MALDI-TOF mass spectrometry was used to identify fragments. Fragments generated by cleavage of peptide bonds immediately before or after the NH2 terminus of the wild-type (WT) and mutant (MT) epitopes are shown. Asterisks indicate unique cleavage sites detected only in the peptide with M1723L substitution by MALDI-TOF mass spectrometry. (B) Wild-type peptide or peptides with M1723L substitution were mock digested in buffer alone (0 h) or buffer containing constitutive proteosomes or immunoproteasomes for 4, 8, or 16 h. Aliquots of the processed peptides (equivalent to approximately 60 μM of the wild-type or mutant substrate) were used to sensitize target cells for lysis by the NS4B1723-specific CTL line. CTL activity was measured at a 20:1 E:T ratio in a 4-h assay. The percentage of specific lysis of target cells sensitized with the mock-digested wild-type or mutant peptides was normalized to 0, and the fold increase or decrease in cytolytic activity with the immunoproteasome-digested peptides was calculated. The percentages of lysis of targets sensitized with mock-digested wild-type and mutant polypeptides were 20 and 35%, respectively. Four percent lysis was detected against unsensitized targets. (C) Wild-type peptides or peptides with M1723L substitution were processed with the immunoproteasome for 16 h as described above, and serial dilutions of the product, equivalent to 20, 10, and 5 μM concentrations of the starting substrate, were used to sensitize target cells. The fold change in cytolytic activity after immunoproteasome treatment was calculated as described above. The percentages of lysis of targets treated with mock-digested wild-type and mutant polypeptide were 22 and 50%, respectively. (D) Wild-type or mutant NS4b1723 peptides were mock digested or treated with the immunoproteasome for 16 h. BLCL cells sensitized with the processed products were cocultured with the NS4B1723-specific CTL line. The frequency of T cells producing IFN-γ was measured by flow cytometry.
Similar articles
- HCV persistence and immune evasion in the absence of memory T cell help.
Grakoui A, Shoukry NH, Woollard DJ, Han JH, Hanson HL, Ghrayeb J, Murthy KK, Rice CM, Walker CM. Grakoui A, et al. Science. 2003 Oct 24;302(5645):659-62. doi: 10.1126/science.1088774. Science. 2003. PMID: 14576438 - The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes.
Erickson AL, Kimura Y, Igarashi S, Eichelberger J, Houghton M, Sidney J, McKinney D, Sette A, Hughes AL, Walker CM. Erickson AL, et al. Immunity. 2001 Dec;15(6):883-95. doi: 10.1016/s1074-7613(01)00245-x. Immunity. 2001. PMID: 11754811 - Positive selection of cytotoxic T lymphocyte escape variants during acute hepatitis C virus infection.
Guglietta S, Garbuglia AR, Pacciani V, Scottà C, Perrone MP, Laurenti L, Spada E, Mele A, Capobianchi MR, Taliani G, Folgori A, Vitelli A, Ruggeri L, Nicosia A, Piccolella E, Del Porto P. Guglietta S, et al. Eur J Immunol. 2005 Sep;35(9):2627-37. doi: 10.1002/eji.200526067. Eur J Immunol. 2005. PMID: 16114108 - Mutational escape from CD8+ T cell immunity: HCV evolution, from chimpanzees to man.
Bowen DG, Walker CM. Bowen DG, et al. J Exp Med. 2005 Jun 6;201(11):1709-14. doi: 10.1084/jem.20050808. J Exp Med. 2005. PMID: 15939787 Free PMC article. Review. - Association of cytotoxic T lymphocyte (CTL) escape mutations with persistent hepatitis C virus (HCV) infection.
Weiner AJ, Erickson AL, Kansopon J, Crawford K, Muchmore E, Houghton M, Walker CM. Weiner AJ, et al. Princess Takamatsu Symp. 1995;25:227-35. Princess Takamatsu Symp. 1995. PMID: 8875628 Review.
Cited by
- Host and viral factors contributing to CD8+ T cell failure in hepatitis C virus infection.
Neumann-Haefelin C, Spangenberg HC, Blum HE, Thimme R. Neumann-Haefelin C, et al. World J Gastroenterol. 2007 Sep 28;13(36):4839-47. doi: 10.3748/wjg.v13.i36.4839. World J Gastroenterol. 2007. PMID: 17828815 Free PMC article. Review. - High-programmed death-1 levels on hepatitis C virus-specific T cells during acute infection are associated with viral persistence and require preservation of cognate antigen during chronic infection.
Rutebemberwa A, Ray SC, Astemborski J, Levine J, Liu L, Dowd KA, Clute S, Wang C, Korman A, Sette A, Sidney J, Pardoll DM, Cox AL. Rutebemberwa A, et al. J Immunol. 2008 Dec 15;181(12):8215-25. doi: 10.4049/jimmunol.181.12.8215. J Immunol. 2008. PMID: 19050238 Free PMC article. - Presence of HCV RNA in peripheral blood mononuclear cells may predict patients response to interferon and ribavirin therapy.
Zaman N, Asad MJ, Raza A, Raja GK, Akhter S, Mahmood M, Mahmood RT. Zaman N, et al. Ann Saudi Med. 2014 Sep-Oct;34(5):401-6. doi: 10.5144/0256-4947.2014.401. Ann Saudi Med. 2014. PMID: 25827697 Free PMC article. - Mutational escape of CD8+ T cell epitopes: implications for prevention and therapy of persistent hepatitis virus infections.
Timm J, Walker CM. Timm J, et al. Med Microbiol Immunol. 2015 Feb;204(1):29-38. doi: 10.1007/s00430-014-0372-z. Epub 2014 Dec 24. Med Microbiol Immunol. 2015. PMID: 25537849 Free PMC article. Review. - Distinct Escape Pathway by Hepatitis C Virus Genotype 1a from a Dominant CD8+ T Cell Response by Selection of Altered Epitope Processing.
Walker A, Skibbe K, Steinmann E, Pfaender S, Kuntzen T, Megger DA, Groten S, Sitek B, Lauer GM, Kim AY, Pietschmann T, Allen TM, Timm J. Walker A, et al. J Virol. 2015 Oct 7;90(1):33-42. doi: 10.1128/JVI.01993-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26446603 Free PMC article.
References
- Appay, V., P. R. Dunbar, M. Callan, P. Klenerman, G. M. Gillespie, L. Papagno, G. S. Ogg, A. King, F. Lechner, C. A. Spina, S. Little, D. V. Havlir, D. D. Richman, N. Gruener, G. Pape, A. Waters, P. Easterbrook, M. Salio, V. Cerundolo, A. J. McMichael, and S. L. Rowland-Jones. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 8:379-385. - PubMed
- Beekman, N. J., P. A. van Veelen, T. van Hall, A. Neisig, A. Sijts, M. Camps, P. M. Kloetzel, J. J. Neefjes, C. J. Melief, and F. Ossendorp. 2000. Abrogation of CTL epitope processing by single amino acid substitution flanking the C-terminal proteasome cleavage site. J. Immunol. 164:1898-1905. - PubMed
- Cardozo, C., and R. A. Kohanski. 1998. Altered properties of the branched chain amino acid-preferring activity contribute to increased cleavages after branched chain residues by the “immunoproteasome.” J. Biol. Chem. 273:16764-16770. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials