Evolutionary origins of genomic repertoires in bacteria - PubMed (original) (raw)
Evolutionary origins of genomic repertoires in bacteria
Emmanuelle Lerat et al. PLoS Biol. 2005 May.
Abstract
Explaining the diversity of gene repertoires has been a major problem in modern evolutionary biology. In eukaryotes, this diversity is believed to result mainly from gene duplication and loss, but in prokaryotes, lateral gene transfer (LGT) can also contribute substantially to genome contents. To determine the histories of gene inventories, we conducted an exhaustive analysis of gene phylogenies for all gene families in a widely sampled group, the gamma-Proteobacteria. We show that, although these bacterial genomes display striking differences in gene repertoires, most gene families having representatives in several species have congruent histories. Other than the few vast multigene families, gene duplication has contributed relatively little to the contents of these genomes; instead, LGT, over time, provides most of the diversity in genomic repertoires. Most such acquired genes are lost, but the majority of those that persist in genomes are transmitted strictly vertically. Although our analyses are limited to the gamma-Proteobacteria, these results resolve a long-standing paradox-i.e., the ability to make robust phylogenetic inferences in light of substantial LGT.
Figures
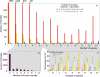
Figure 1. Distribution of Gene Families and Occurrence of LGT in γ-Proteobacteria
(A) Numbers of species and numbers of synologs corresponding to the 14,158 gene families. Single-copy gene families (red bars) comprise the large majority of the families. The numbers of families in categories exceeding 300 members are displayed on top. (B) Losses required to reconcile gene distribution with organismal phylogeny [16] for gene families represented in fewer than six species. For each family, we inferred an initial acquisition event in the most recent ancestor of the species containing a gene from the family and tallied the minimum number of independent events of loss required to explain the phylogenetic distribution. Most distributions can be explained without invoking multiple gene losses, supporting the hypothesis of a single acquisition. (C) Percentage of families containing fewer than three synologs (# gene copies − # genomes = 0, 1, or 2) showing evidence of LGT by the method described in Lerat et al. [16] and Figure 2. Boxes represent the conservative estimate of LGT and dashed bars represent the corresponding permissive estimate (see text). For families containing additional synologs (white bars), it was not practical to apply the same method; instead, we built neighbor-joining trees (see Materials and Methods).
Figure 2. Testing for LGT and Duplication as Sources of Synology
The illustrated case is for families with a single synolog, that is, in which one genome contains two gene copies. We tested two alignments, each retaining one of the two copies (red or blue), against the reference organismal phylogeny [16]. When both alignments agreed with the reference tree (+/+), the synology could be attributed to recent intragenomic duplication, whereas in cases of phylogenetic incongruence of one of the alignments (+/− or −/+), LGT of one synolog was invoked. If both alignments rejected the reference tree (−/−), the family was considered as containing one or several LGT events. Tests of LGT were conducted similarly for families with an additional synolog (one genome with three synologs or two genomes each with two synologs). In such cases, each possible alignment containing a single copy per genome was tested. In addition to these tests, all family trees were inspected to confirm diagnoses of LGT.
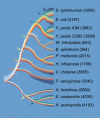
Figure 3. LGT and Genome Evolution in γ-Proteobacteria
Only a small proportion of genes have been retained since the common ancestor of γ-proteobacteria (in red). Under the assumption that ancestral and contemporary genome sizes are similar, most of the genes present in this ancestral genome (in white) have been replaced by nonhomologous genes (yellow to green), usually via LGT from organisms outside of this clade. Once a new gene is acquired, its transmission follows vertical inheritance. The abundance of genes unique to a species (in blue) indicates that these bacteria (with the exception of the endosymbionts) constantly acquire new genes, most of which do not persist long-term within lineages. (Numbers of protein-coding genes, excluding those corresponding to known IS elements and phages, are in parentheses for each genome).
Similar articles
- From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-Proteobacteria.
Lerat E, Daubin V, Moran NA. Lerat E, et al. PLoS Biol. 2003 Oct;1(1):E19. doi: 10.1371/journal.pbio.0000019. Epub 2003 Sep 15. PLoS Biol. 2003. PMID: 12975657 Free PMC article. - The evolutionary history of quorum-sensing systems in bacteria.
Lerat E, Moran NA. Lerat E, et al. Mol Biol Evol. 2004 May;21(5):903-13. doi: 10.1093/molbev/msh097. Epub 2004 Mar 10. Mol Biol Evol. 2004. PMID: 15014168 - Evolution of glutamate dehydrogenase genes: evidence for lateral gene transfer within and between prokaryotes and eukaryotes.
Andersson JO, Roger AJ. Andersson JO, et al. BMC Evol Biol. 2003 Jun 23;3:14. doi: 10.1186/1471-2148-3-14. Epub 2003 Jun 23. BMC Evol Biol. 2003. PMID: 12820901 Free PMC article. - Phylogenomic networks.
Dagan T. Dagan T. Trends Microbiol. 2011 Oct;19(10):483-91. doi: 10.1016/j.tim.2011.07.001. Epub 2011 Aug 3. Trends Microbiol. 2011. PMID: 21820313 Review.
Cited by
- Identification of homologs of the Chlamydia trachomatis effector CteG reveals a family of Chlamydiaceae type III secreted proteins that can be delivered into host cells.
Pereira IS, da Cunha M, Leal IP, Luís MP, Gonçalves P, Gonçalves C, Mota LJ. Pereira IS, et al. Med Microbiol Immunol. 2024 Jul 15;213(1):15. doi: 10.1007/s00430-024-00798-9. Med Microbiol Immunol. 2024. PMID: 39008129 Free PMC article. - Epidemiological exploration of fleas and molecular identification of flea-borne viruses in Egyptian small ruminants.
Barghash SM, Yassin SE, Sadek AM, Mahmoud DM, Salama MS. Barghash SM, et al. Sci Rep. 2024 Jul 2;14(1):15166. doi: 10.1038/s41598-024-64881-0. Sci Rep. 2024. PMID: 38956077 Free PMC article. - Parameter Estimation and Species Tree Rooting Using ALE and GeneRax.
Williams TA, Davín AA, Morel B, Szánthó LL, Spang A, Stamatakis A, Hugenholtz P, Szöllősi GJ. Williams TA, et al. Genome Biol Evol. 2023 Jul 3;15(7):evad134. doi: 10.1093/gbe/evad134. Genome Biol Evol. 2023. PMID: 37463417 Free PMC article. - Conjugative plasmids inhibit extracellular electron transfer in Geobacter sulfurreducens.
Fessler M, Madsen JS, Zhang Y. Fessler M, et al. Front Microbiol. 2023 Mar 17;14:1150091. doi: 10.3389/fmicb.2023.1150091. eCollection 2023. Front Microbiol. 2023. PMID: 37007462 Free PMC article. - In silico analysis reveals the co-existence of CRISPR-Cas type I-F1 and type I-F2 systems and its association with restricted phage invasion in Acinetobacter baumannii.
Yadav G, Singh R. Yadav G, et al. Front Microbiol. 2022 Aug 17;13:909886. doi: 10.3389/fmicb.2022.909886. eCollection 2022. Front Microbiol. 2022. PMID: 36060733 Free PMC article.
References
- Ohno S. Evolution by gene duplication. Heidelberg (Germany): Springer-Verlag; 1970. 160 pp.
- Ohta T. Role of gene duplication in evolution. Genome. 1989;31:304–310. - PubMed
- Zhang JZ. Evolution by gene duplication: An update. Trends Ecol Evol. 2003;18:292–298.
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources