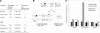Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress - PubMed (original) (raw)
Comparative Study
. 2005 Apr 15;19(8):919-32.
doi: 10.1101/gad.1304305. Epub 2005 Apr 1.
Affiliations
- PMID: 15805465
- PMCID: PMC1080131
- DOI: 10.1101/gad.1304305
Comparative Study
Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress
Mihoko Kai et al. Genes Dev. 2005.
Abstract
The replication checkpoint kinase Cds1 preserves genome integrity by stabilizing stalled replication forks. Cds1 targets substrates through its FHA domain. The Cds1 FHA domain interacts with Mus81, a subunit of the Mus81-Eme1 structure-specific endonuclease. We report here that Mus81 and Rhp51 are required for generating deletion mutations in fission yeast replication mutants that experience replication stress. A mutation in the Mus81 FHA-binding motif eliminates its Cds1-binding and Cds1-dependent phosphorylation. Furthermore, this mutation exacerbates the deletion mutator phenotype of a replication mutant, and induces a hyper-recombination phenotype in hydroxyurea-treated cells. In unperturbed cells, Mus81 associates with chromatin throughout S phase. In replication mutants grown at semipermissive temperature, Mus81 undergoes minor Cds1-dependent phosphorylation, remains chromatin-associated, generates deletion mutations, and maintains cell growth. Upon S-phase arrest by acute hydroxyurea treatment, Mus81 is not required for cell viability but is essential for recovery from replication fork collapse. Moreover, Mus81 undergoes extensive Cds1-dependent phosphorylation and dissociates from chromatin in hydroxyurea-arrested cells, thereby preventing it from cleaving stalled replication forks that could lead to fork breakage and chromosomal rearrangement. These results provide novel insights into how Cds1 regulates Mus81 accordingly when cells experience different replication stress to preserve genome integrity.
Figures
Figure 1.
Mutations in pol_α(swi7-H4)_ cells with mus81, eme1, and rhp51 mutant backgrounds. (A) Mutation rates. Mutation rates are the average of three or more experiments. C.I. is the 95% confidence interval (10-8 per cell division). Fold increases or decreases in mutation rates relative to the pol_α(swi7-H4)_ single mutant are shown as ↑ or ↓, respectively. (B) Types of mutations. The rates of each mutation type were calculated by multiplying the rate of 5-FOA survival cells with the percentage of alterations that are shown as deletion or point mutations. “Point mutation” represents both base substitutions and single-base frameshift mutations. Bar graphs show the types of mutation rate relative to wild type.
Figure 2.
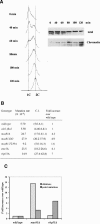
A mutation in Mus81 T-X-X-D motif abolishes the Mus81 protein association and phosphorylation by Cds1. (A) Mapping the Cds1-binding site on Mus81. Five overlapping fragments of mus81+ encoding Mus81 from residue 1 to 314 were constructed as a GST-fusion. The GST-fusion Mus81 proteins were expressed from bacteria, purified, and used as affinity matrix for binding of myc-tagged Cds1 from fission yeast cell extracts. (B) Mutation in mus81.T239A abolishes the binding of Mus81 with Cds1. A GST-fusion protein containing the 1-190 region of Cds1, which contains the FHA domain, was overexpressed in strains that expressed 13-myc-tagged forms of wild type or mus81.T239A from the mus81 genomic locus. GST-Cds1-fusion protein was purified with GST-Sepharose and probed for coprecipitating Mus81:13myc. (C) Mutation in mus81.T239A abolishes the HU-induced phosphorylation of Mus81. Strains that expressed 13-myc-tagged forms of wild type or mus81.T239A at endogenous levels were treated with 10 mM HU for 4 h. Mus81 protein was detected by immunoblotting with anti-myc antibody. Wild-type but not mutant Mus81 displayed reduced electrophoretic mobility, indicative of Cds1-dependent phosphorylation (Boddy et al. 2000).
Figure 3.
Mutation in mus81.T239A or cds1.fha1 enhances the deletion mutator phenotype in pol_α(swi7-H4). (A) Mutation rates. Mutation in mus81.T239A or cds1.fha1 enhances the deletion mutation rate in a pol_α(swi7-H4) background. Mutation rates were measured as described in Materials and Methods and Figure 1. (B) Mutation types. Rates of deletion mutation and point mutation of each strain relative to wild type are shown as bar graphs and calculated as described in Figure 1.
Figure 4.
Cds1-dependent phosphorylation of Mus81 promotes Mus81 to dissociate from chromatin. (A) Replication stress induces Mus81 phosphorylation and chromatin dissociation. Logarithmically growing wild-type cells harboring mus81+:myc were treated with 12 mM HU for 3 h at 30°C. pol_α(swi7-H4):mus81_+:myc cells were grown at 30°C and 34°C to induce replication stress. The chromatin fractionation assay was performed as described (Kai and Wang 2003b). (B) HU-induced chromatin dissociation of Mus81 is impaired in mus81.T239A and cds1.fha1 cells. The chromatin fractionation assay was performed with logarithmically growing mus81.T239:myc and cds1.fha1 harboring mus81+:myc treated with 12 mM HU for 3 h at 30°C as described above. (C) Mus81 protein is mildly phosphorylated and remains chromatin-associated in CPT-treated cells. The chromatin fractionation assay was performed with logarithmically growing wild-type cells treated with 30 μM of CPT for 2 h at 30°C. (D) Chromatin fractionation assay control. Histone H4 and α-tubulin were used as controls for chromatin-bound and non-chromatin-bound proteins, respectively, in either hydroxyurea-treated cells, in pol_α(swi7-H4)_ mutant, CPT-treated cells, or untreated control wild-type cells by immunoblotting with anti-histone H4 polyclonal antibody or an anti-tubulin monoclonal antibody.
Figure 5.
Mus81 is not required to survive HU-induced stalled replication, but is required for surviving replication fork collapse. Logarithmically growing wild-type and mus81_Δ cells were incubated at 30°C with either 12 mM HU or 30 μM CPT. (A) mus81_Δ cells are not sensitive to acute HU treatment but sensitive to CPT treatment. Cells incubated in HU or CPT were removed in every 2 h and assay for viability. (B) CPT treatment does not activate Cds1 kinase activity in cells. Wild-type cells were incubated in either 12 mM HU or 30 μM CPT for 4 h. Cell extracts were prepared and Cds1 kinase activity was analyzed as described (Lindsay et al. 1998) with myelin basic protein (MBP) as substrate. (C) Mus81 protein is mildly phosphorylated in CPT-treated cells. Wild-type cells containing mus81+:myc were treated with either HU or CPT as described above. Cell extracts were fractionated by gel electrophoresis followed by immunoblotting with anti-myc antibody.
Figure 6.
HU treatment of mus81.T239A mutant enhances the cells' recombination frequency. Logarithmically growing wild-type, mus81.T239A, and _mus81_Δ cells were incubated with or without 12 mM HU for 5 h and assayed for recombination frequency and recombination types as described in Materials and Methods. (A) Spontaneous and HU-induced recombination frequencies. (B) Schematic of the recombination substrate and recombination products. Solid and open circles represent the ade6-L469 and ade6-M375 mutations, respectively. (C) The mus81.T239A cells undergo a conversion type of recombination in response to acute HU treatment.
Figure 7.
Mus81 has a role in maintaining genome integrity during the normal cell cycle. (A) Cells were synchronized by cdc10-m17 arrest in G1 at 36.5°C for 4.5 h and released to 25°C. Cell samples were collected at G1 arrest and every 20 min after release from G1 arrest. Cell cycle progression is shown as FACS profile (left panel) and chromatin association of Mus81 protein was shown (right panel). (B) The mutation rate of _cds1.fha1, mus81_Δ, _mus81.DD, mus81.T239A, eme1_Δ, and _rhp51_Δ cells without replication stress. Mutation rates were calculated as described in Materials and Methods and Figure 1. (C) Types of mutations in _mus81_Δ and _rhp51_Δ cells without replication stress. Mutation types were calculated as described in Figure 1. Bar graphs show the fold of deletion and point mutations in unperturbed _mus81_Δ and _rhp51_Δ cells over wild-type cells.
Figure 8.
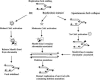
A model of how Cds1 regulates Mus81-Eme1 in response to replication stress. Replication stalling caused by HU arrest robustly activates Cds1 kinase. The robust Cds1-dependent phosphorylation of Mus81 promotes Mus81-Eme1 to dissociate from chromatin, thus preventing Mus81-Eme1 from cleaving the stalled replication forks and stabilizing the fork, resulting in no deletion mutation found in HU-arrested cells. This is a strategy of cells to survive HU-induced replication stalling. Replication stress caused by a replication mutant induces a moderate level of Cds1 kinase activation. A moderate Cds1-dependent phosphorylation of Mus81 allows Mus81 to remain chromatin-associated to generate some deletion of genomic sequences to tolerate the replication stress, to facilitate replication fork restart, and to maintain an active cell growth. Cds1 is not activated by spontaneous replication fork collapse, and Mus81 remains chromatin-associated. The chromatin-associated Mus81 could also generate deletion mutations in a replication mutant.
Similar articles
- Cleavage of stalled forks by fission yeast Mus81/Eme1 in absence of DNA replication checkpoint.
Froget B, Blaisonneau J, Lambert S, Baldacci G. Froget B, et al. Mol Biol Cell. 2008 Feb;19(2):445-56. doi: 10.1091/mbc.e07-07-0728. Epub 2007 Nov 21. Mol Biol Cell. 2008. PMID: 18032583 Free PMC article. - Active Replication Checkpoint Drives Genome Instability in Fission Yeast mcm4 Mutant.
Kim SM, Forsburg SL. Kim SM, et al. Mol Cell Biol. 2020 Jun 29;40(14):e00033-20. doi: 10.1128/MCB.00033-20. Print 2020 Jun 29. Mol Cell Biol. 2020. PMID: 32341083 Free PMC article. - Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1.
Noguchi E, Noguchi C, Du LL, Russell P. Noguchi E, et al. Mol Cell Biol. 2003 Nov;23(21):7861-74. doi: 10.1128/MCB.23.21.7861-7874.2003. Mol Cell Biol. 2003. PMID: 14560029 Free PMC article. - Checkpoint responses to replication stalling: inducing tolerance and preventing mutagenesis.
Kai M, Wang TS. Kai M, et al. Mutat Res. 2003 Nov 27;532(1-2):59-73. doi: 10.1016/j.mrfmmm.2003.08.010. Mutat Res. 2003. PMID: 14643429 Review. - Checking in on Cds1 (Chk2): A checkpoint kinase and tumor suppressor.
McGowan CH. McGowan CH. Bioessays. 2002 Jun;24(6):502-11. doi: 10.1002/bies.10101. Bioessays. 2002. PMID: 12111733 Review.
Cited by
- Schizosaccharomyces pombe Cds1Chk2 regulates homologous recombination at stalled replication forks through the phosphorylation of recombination protein Rad60.
Miyabe I, Morishita T, Shinagawa H, Carr AM. Miyabe I, et al. J Cell Sci. 2009 Oct 15;122(Pt 20):3638-43. doi: 10.1242/jcs.046508. Epub 2009 Sep 15. J Cell Sci. 2009. PMID: 19755492 Free PMC article. - Essential domains of Schizosaccharomyces pombe Rad8 required for DNA damage response.
Ding L, Forsburg SL. Ding L, et al. G3 (Bethesda). 2014 May 28;4(8):1373-84. doi: 10.1534/g3.114.011346. G3 (Bethesda). 2014. PMID: 24875629 Free PMC article. - Control of structure-specific endonucleases to maintain genome stability.
Dehé PM, Gaillard PHL. Dehé PM, et al. Nat Rev Mol Cell Biol. 2017 May;18(5):315-330. doi: 10.1038/nrm.2016.177. Epub 2017 Mar 22. Nat Rev Mol Cell Biol. 2017. PMID: 28327556 Review. - Mus81, Rhp51(Rad51), and Rqh1 form an epistatic pathway required for the S-phase DNA damage checkpoint.
Willis N, Rhind N. Willis N, et al. Mol Biol Cell. 2009 Feb;20(3):819-33. doi: 10.1091/mbc.e08-08-0798. Epub 2008 Nov 26. Mol Biol Cell. 2009. PMID: 19037101 Free PMC article. - The intra-S phase checkpoint directly regulates replication elongation to preserve the integrity of stalled replisomes.
Liu Y, Wang L, Xu X, Yuan Y, Zhang B, Li Z, Xie Y, Yan R, Zheng Z, Ji J, Murray JM, Carr AM, Kong D. Liu Y, et al. Proc Natl Acad Sci U S A. 2021 Jun 15;118(24):e2019183118. doi: 10.1073/pnas.2019183118. Proc Natl Acad Sci U S A. 2021. PMID: 34108240 Free PMC article.
References
- Bahler J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie III, A., Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943-951. - PubMed
- Boddy M.N. and Russell, P. 2001. DNA replication checkpoint. Curr. Biol. 11: R953-R956. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases