Calbindin D28k targets myo-inositol monophosphatase in spines and dendrites of cerebellar Purkinje neurons - PubMed (original) (raw)
Calbindin D28k targets myo-inositol monophosphatase in spines and dendrites of cerebellar Purkinje neurons
Hartmut Schmidt et al. Proc Natl Acad Sci U S A. 2005.
Abstract
The Ca(2+)-binding protein calbindin D28k (CB) is vital for the normal function of the central nervous system but its specific functional role is largely unclear. CB is typically described as a mobile Ca(2+)buffer that shapes the spatiotemporal extent of cellular Ca(2+)signals. Recent biochemical data, however, indicate that CB also has characteristics of a Ca(2+) sensor and activates myo-inositol monophosphatase (IMPase), a key enzyme of the inositol-1,4,5-trisphosphate signaling cascade and an assumed target of mood-stabilizing drugs in the treatment of bipolar disorder. Here, we show that CB interacts with IMPase in cerebellar Purkinje neurons, a cell type well known to rely on inositol-1,4,5-trisphosphate-dependent synaptic integration. Quantification of the mobility of dye-labeled CB with two-photon fluorescence recovery after photobleaching revealed that a substantial fraction of CB is immobilized in spines and dendrites, but not in axons. Immobilization occurs over several seconds, is increased by suprathreshold synaptic activity, and can be relieved by a synthetic peptide that resembles the putative CB-binding site of IMPase, indicating that CB binds to immobilized IMPase. Measurements of the apparent diffusion coefficients of CB imply that CB does not interact with cytosolic IMPase or that the latter is present only in minute amounts in the spiny dendrites of Purkinje neurons. Our results suggest that CB acts as an activity-dependent sensor that targets membrane/cytoskeleton-bound IMPase in central neurons.
Figures
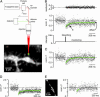
Fig. 1.
CB* is partly immobilized in spines and dendrites but not in axons of cerebellar PNs. (A) Experimental set-up for FRAP recordings. Mode-locked laser light was intensity-modulated (Icmd) by a Pockels cell and scanned with a custom-modified laser scanning microscope. The image shows spiny dendrites of a PN loaded with 200 μM CB* via a somatic patch pipette. For FRAP recordings the laser beam was focused on a single point of interest. (B) FRAP recording from the spine illustrated in A. The recovery (Middle) could be described by a single exponential function (green line; τ = 81 ms) with an offset that indicates that 31% of CB* was immobilized in the spine. Shown are the residuals for the fit (Top) and the laser intensity (Bottom), respectively. (C) FRAP recording from a spine labeled with 40-kDa fluorescein dextran. Note the complete return to baseline. (D and E) FRAP of CB* in a dendritic (D) and an axonal (E) compartment of a PN. Note the presence and absence, respectively, of the offset.
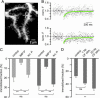
Fig. 2.
CB* is immobilized by IMPase. (A) Image of spiny dendrites codialyzed with CB* and IMP-P, a synthetic peptide identical to the putative CB binding site of IMPase-1 (7). (B) FRAP recordings from the spine (Upper) and the parent dendrite (Lower) marked by crosshairs in A. Note the almost complete return to baseline. (C) Average immobilization of CB* in spines (Left) and dendrites (Right) under control conditions and in the presence of IMP-P or the scrambled control peptide (scr. P). (D) Immobilization of CB* in dendrites during ongoing synaptic stimulation (CF stim.). **, P ≤ 0.001; ns, not significant.
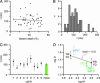
Fig. 3.
Diffusional mobility of CB*. (A) Plot of FRAP time constants vs. bleach depth (n = 49 spine recordings, r =–0,07, solid line). (B) Distribution of τ values. (C) Variability of τ values within (median and interquartile range) and between cells (mean ± SE of medians). (D) Double-logarithmic plot of the derived apparent diffusion coefficients (D) of CB* in the absence (light green) and the presence (dark green) of IMP-P plotted against their molecular masses. The IMP-P was assumed to form (1:1) dimers with CB*. Published D values of 10- and 40-kDa fluorescein dextran (FD) and dye-labeled PV (PV*) are also shown (25). The line represents a linear fit to the FD data.
Fig. 4.
Slow unbinding of CB* from IMPase. (A) CB*-labeled spiny dendrites. (B) Long-lasting, intermittent FRAP recording in the spine marked in A. The recovery could be described by the sum of two exponential functions (green line). The slow time constant (τslow) was 4.7 s. (C) Average τslow values (median ± interquartile range) from 10 spines from five cells.
Similar articles
- Myo-inositol monophosphatase is an activated target of calbindin D28k.
Berggard T, Szczepankiewicz O, Thulin E, Linse S. Berggard T, et al. J Biol Chem. 2002 Nov 1;277(44):41954-9. doi: 10.1074/jbc.M203492200. Epub 2002 Aug 9. J Biol Chem. 2002. PMID: 12176979 - Interaction of calbindin D28k and inositol monophosphatase in human postmortem cortex: possible implications for bipolar disorder.
Shamir A, Elhadad N, Belmaker RH, Agam G. Shamir A, et al. Bipolar Disord. 2005 Feb;7(1):42-8. doi: 10.1111/j.1399-5618.2004.00162.x. Bipolar Disord. 2005. PMID: 15654931 - Bergmann glial S100B activates myo-inositol monophosphatase 1 and Co-localizes to purkinje cell vacuoles in SCA1 transgenic mice.
Vig PJ, Shao Q, Subramony SH, Lopez ME, Safaya E. Vig PJ, et al. Cerebellum. 2009 Sep;8(3):231-44. doi: 10.1007/s12311-009-0125-5. Epub 2009 Jul 11. Cerebellum. 2009. PMID: 19593677 Free PMC article. - Emerging role of inositol monophosphatase in cancer.
Chen Q, Shen L, Li S. Chen Q, et al. Biomed Pharmacother. 2023 May;161:114442. doi: 10.1016/j.biopha.2023.114442. Epub 2023 Feb 24. Biomed Pharmacother. 2023. PMID: 36841024 Review.
Cited by
- Harnessing the Power of Purple Sweet Potato Color and _Myo_-Inositol to Treat Classic Galactosemia.
Hagen-Lillevik S, Johnson J, Siddiqi A, Persinger J, Hale G, Lai K. Hagen-Lillevik S, et al. Int J Mol Sci. 2022 Aug 4;23(15):8654. doi: 10.3390/ijms23158654. Int J Mol Sci. 2022. PMID: 35955788 Free PMC article. - Ca(2+) sensor proteins in dendritic spines: a race for Ca(2+).
Raghuram V, Sharma Y, Kreutz MR. Raghuram V, et al. Front Mol Neurosci. 2012 May 8;5:61. doi: 10.3389/fnmol.2012.00061. eCollection 2012. Front Mol Neurosci. 2012. PMID: 22586368 Free PMC article. - Regulation of renal calbindin expression during cisplatin-induced kidney injury.
George B, Szilagyi JT, Joy MS, Aleksunes LM. George B, et al. J Biochem Mol Toxicol. 2022 Jul;36(7):e23068. doi: 10.1002/jbt.23068. Epub 2022 Apr 10. J Biochem Mol Toxicol. 2022. PMID: 35403300 Free PMC article. - Distribution Patterns of Subgroups of Inhibitory Neurons Divided by Calbindin 1.
Zhang B, Li L, Tang X, Zeng J, Song Y, Hou Z, Ma T, Afewerky HK, Li H, Lu Y, He A, Li X. Zhang B, et al. Mol Neurobiol. 2023 Dec;60(12):7285-7296. doi: 10.1007/s12035-023-03542-1. Epub 2023 Aug 7. Mol Neurobiol. 2023. PMID: 37548854 - Three functional facets of calbindin D-28k.
Schmidt H. Schmidt H. Front Mol Neurosci. 2012 Mar 15;5:25. doi: 10.3389/fnmol.2012.00025. eCollection 2012. Front Mol Neurosci. 2012. PMID: 22435048 Free PMC article.
References
- Baimbridge, K. G., Celio, M. R. & Rogers, J. H. (1992) Trends Neurosci. 15, 303–308. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous