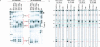The contributions of dsRNA structure to Dicer specificity and efficiency - PubMed (original) (raw)
The contributions of dsRNA structure to Dicer specificity and efficiency
Annaleen Vermeulen et al. RNA. 2005 May.
Abstract
Dicer processes long double-stranded RNA (dsRNA) and pre-microRNAs to generate the functional intermediates (short interfering RNAs and microRNAs) of the RNA interference pathway. Here we identify features of RNA structure that affect Dicer specificity and efficiency. The data presented show that various attributes of the 3' end structure, including overhang length and sequence composition, play a primary role in determining the position of Dicer cleavage in both dsRNA and unimolecular, short hairpin RNA (shRNA). We also demonstrate that siRNA end structure affects overall silencing functionality. Awareness of these new features of Dicer cleavage specificity as it is related to siRNA functionality provides a more detailed understanding of the RNAi mechanism and can shape the development of hairpins with enhanced functionality.
Figures
FIGURE 1.
61 mer MAPK14 dsRNA containing blunt and overhang ends are processed in distinctly different manners. (A) Dicer cleavage of [32P]-5′ sense labeled dsRNA with two blunt ends (blunt–blunt) and two 2-nt overhang ends (over–over) analyzed by denaturing PAGE. Incubation times in minutes. RNase T1 digest (T1) and hydroxyl digest (OH) are labeled at the tops of the lanes for both substrates. The primary and secondary cleavage product bands that result from Dicer entering from the 5′ sense (left) and 3′ sense (right) ends are indicated in red and black, respectively. (B) Diagrams of Dicer processing of blunt and/or overhang ended MAPK14 61mer dsRNA substrates. Red and black arrows show Dicer positions of cleavage from the 5′ sense (left) and 3′ sense (right) ends, respectively. Solid arrows were derived from Dicer digestion of [32P]-5′ sense or antisense labeled dsRNA. Length of arrows indicates intensity of cleavage. Processing positions with dashed arrows were derived from processing of blunt secondary products (Supplementary Fig. 1 at
www.dharmacon.com/tech/publications
).
FIGURE 2.
End structure and sequence composition determine Dicer cleavage pattern. (A) Processing of MAPK14 61mer dsRNA containing blunt and/or 2-nt 3′ overhang ends by Dicer analyzed by denaturing PAGE. Lanes are labeled describing dsRNA termini oriented relative to sense strand from 5′ to 3′. Sense or antisense labeled strands are shown on the left and right, respectively. (P) 5′ end of terminus was 5′ (nonradio-actively) phosphorylated during synthesis. (B) The contribution of sequence to Dicer cleavage specificity. MAPK14, CYCLO, and DBI 61mer sequences containing (1) blunt ends (blunt–blunt) or (2) 2-nt 3′ overhang ends (over–over) were digested by Dicer. Both (1) sense and (2) antisense labeled duplexes are shown. Lanes from separate gels were sized to enable side-by-side comparison of bands for different digests. (Original gels are in Supplementary Figs. 2–4 at
www.dharmacon.com/tech/publications
.)
FIGURE 3.
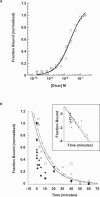
Long dsRNA 61mers containing blunt or overhang ends have the same affinity to Dicer holoenzyme. (A) Determination of dissociation constant (K d) of Dicer–RNA complex by nitrocellulose binding assay. Fraction [32P] RNA duplex bound versus Dicer concentration for 61-nt MAPK14 dsRNA duplex with 2-nt 3′ overhang (○) and blunt (□) ends. (B) Determination of Dicer–RNA complex _t_1/2 by pulse-chase experiment after a 1 min and 30 min preincubation. Fraction [32P]-labeled RNA duplex bound to 80 nM Dicer was measured as a function of time after addition of 100 nM unlabeled dsRNA (chase). One minute preincubation prior to addition of unlabeled chase: 61-nt MAPK14 dsRNA duplex with 2-nt 3′ overhang (▪); 61-nt MAPK14 blunt dsRNA duplex (•). Thirty minute preincubation: 61-nt MAPK14 dsRNA duplex with 2-nt 3′ overhang (□); 61-nt MAPK14 blunt dsRNA duplex (○). (Insert) Log-plot of pulse-chase experiment.
FIGURE 4.
Length and sequence composition of overhangs affect Dicer processing of dsRNA. (A) Dicer cleavage of dsRNA with 2-nt 3′overhang of every possible sequence combination analyzed by denaturing PAGE. Diagram of MAPK14 61mer duplex indicates position of 5′-[32P]-labeling. (XX) position of 3′ overhang nucleotides. Lanes are labeled with sequence 5′-3′. (B) Quantification of primary Dicer cleavage products from the 2-nt 3′overhang end for different sequence combinations. The data are sorted from least to most efficient cleavage. Residues boxed in red result in less efficient cleavage while residues boxed in blue result in more efficient cleavage. The graph shows the average from three experiments and error bars indicate standard deviation. The vertical axis is scaled from 20% to 55% to emphasize differences in cleavage efficiency. (C) Dicer cleavage of dsRNA with 3′ overhang of length of 0–5 nt (natural sequence).
FIGURE 5.
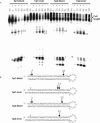
End structure has a strong impact on Dicer processing of shRNA. (A) Dicer processing of two cyclophilin directed shRNAs containing blunt or 2-nt 3′ overhang ends. shRNAs were incubated in the presence (+) and absence (−) of Dicer for varying periods of time (5–60 min). (B) Characteristic cleavage product sizes are indicated (26, 25, and 3 nt for blunt substrates, 23 and 22 nt for overhang substrates). Small gray arrows indicate products resulting from digestion of a small percentage of material having a nicked loop.
FIGURE 6.
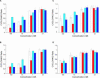
siRNA activity is affected by asymmetric end structure. Silencing efficiency of siRNAs containing blunt and/or overhang ends at different concentrations (over–over [white], over–blunt [red], blunt–over [cyan], and blunt–blunt [blue] for siRNAs 1, 2, 3, and 4 in A,B,C,D, respectively). Silencing is expressed as a ratio of Cyclophilin B and house keeping gene (HKG) mRNA expression.
Similar articles
- Single processing center models for human Dicer and bacterial RNase III.
Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Zhang H, et al. Cell. 2004 Jul 9;118(1):57-68. doi: 10.1016/j.cell.2004.06.017. Cell. 2004. PMID: 15242644 - Homodimeric structure and double-stranded RNA cleavage activity of the C-terminal RNase III domain of human dicer.
Takeshita D, Zenno S, Lee WC, Nagata K, Saigo K, Tanokura M. Takeshita D, et al. J Mol Biol. 2007 Nov 16;374(1):106-20. doi: 10.1016/j.jmb.2007.08.069. Epub 2007 Sep 8. J Mol Biol. 2007. PMID: 17920623 - Continuous fluorescence-based method for assessing dicer cleavage efficiency reveals 3' overhang nucleotide preference.
DiNitto JP, Wang L, Wu JC. DiNitto JP, et al. Biotechniques. 2010 Apr;48(4):303-11. doi: 10.2144/000113402. Biotechniques. 2010. PMID: 20569207 - [RNA binding proteins in the RNA interference phenomenon].
Kotel'nikov RN, Shpiz SG, Kalmykova AI, Gvozdev VA. Kotel'nikov RN, et al. Mol Biol (Mosk). 2006 Jul-Aug;40(4):595-608. Mol Biol (Mosk). 2006. PMID: 16913219 Review. Russian. - [Great potential of small RNAs: RNA interference and microRNA].
Vázquez-Ortiz G, Piña-Sánchez P, Salcedo M. Vázquez-Ortiz G, et al. Rev Invest Clin. 2006 Jul-Aug;58(4):335-49. Rev Invest Clin. 2006. PMID: 17146945 Review. Spanish.
Cited by
- Substrate promiscuity of Dicer toward precursors of the let-7 family and their 3'-end modifications.
Dadhwal G, Samy H, Bouvette J, El-Azzouzi F, Dagenais P, Legault P. Dadhwal G, et al. Cell Mol Life Sci. 2024 Jan 23;81(1):53. doi: 10.1007/s00018-023-05090-2. Cell Mol Life Sci. 2024. PMID: 38261114 Free PMC article. - Design of extended short hairpin RNAs for HIV-1 inhibition.
Liu YP, Haasnoot J, Berkhout B. Liu YP, et al. Nucleic Acids Res. 2007;35(17):5683-93. doi: 10.1093/nar/gkm596. Epub 2007 Aug 21. Nucleic Acids Res. 2007. PMID: 17715143 Free PMC article. - Identification and characterization of Dicer1e, a Dicer1 protein variant, in oral cancer cells.
Cantini LP, Andino LM, Attaway CC, Butler B, Dumitriu A, Blackshaw A, Jakymiw A. Cantini LP, et al. Mol Cancer. 2014 Aug 13;13:190. doi: 10.1186/1476-4598-13-190. Mol Cancer. 2014. PMID: 25115815 Free PMC article. - Design and implementation of a synthetic pre-miR switch for controlling miRNA biogenesis in mammals.
Atanasov J, Groher F, Weigand JE, Suess B. Atanasov J, et al. Nucleic Acids Res. 2017 Dec 15;45(22):e181. doi: 10.1093/nar/gkx858. Nucleic Acids Res. 2017. PMID: 29036355 Free PMC article. - A potential protein-RNA recognition event along the RISC-loading pathway from the structure of A. aeolicus Argonaute with externally bound siRNA.
Yuan YR, Pei Y, Chen HY, Tuschl T, Patel DJ. Yuan YR, et al. Structure. 2006 Oct;14(10):1557-65. doi: 10.1016/j.str.2006.08.009. Structure. 2006. PMID: 17027504 Free PMC article.
References
- Bartel, D. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. - PubMed
- Bogenhagen, D. and Brown, D. 1981. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell 24: 261–270. - PubMed
- Brummelkamp, T.R., Bernards, R., and Agami, R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources