Ipr1 gene mediates innate immunity to tuberculosis - PubMed (original) (raw)
Ipr1 gene mediates innate immunity to tuberculosis
Hui Pan et al. Nature. 2005.
Abstract
An estimated eight million people are infected each year with the pathogen Mycobacterium tuberculosis, and more than two million die annually. Yet only about 10% of those infected develop tuberculosis. Genetic variation within host populations is known to be significant in humans and animals, but the nature of genetic control of host resistance to tuberculosis remains poorly understood. Previously we mapped a new genetic locus on mouse chromosome 1, designated sst1 (for supersusceptibility to tuberculosis 1). Here we show that this locus mediates innate immunity in sst1 congenic mouse strains and identify a candidate gene, Intracellular pathogen resistance 1 (Ipr1), within the sst1 locus. The Ipr1 gene is upregulated in the sst1 resistant macrophages after activation and infection, but it is not expressed in the sst1 susceptible macrophages. Expression of the Ipr1 transgene in the sst1 susceptible macrophages limits the multiplication not only of M. tuberculosis but also of Listeria monocytogenes and switches a cell death pathway of the infected macrophages from necrosis to apoptosis. Our data indicate that the Ipr1 gene product might have a previously undocumented function in integrating signals generated by intracellular pathogens with mechanisms controlling innate immunity, cell death and pathogenesis.
Conflict of interest statement
Competing interests statement The authors declare that they have no competing financial interests.
Figures
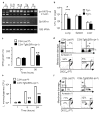
Figure 1. The sst1 locus mediates innate immunity to tuberculosis.
a, b, Survival of C3H, B6, and the C3H.B6-sst1 (sst1 R) mice after i.v. (a) or aerosol (b) infection with MTB; c, MTB bacterial loads in the lungs of the sst1 congenic mice after the aerosol infection; d, e Tuberculosis lung lesions 25 days after i.v. infection (d) and 12 weeks after aerosol infection, H&E, 40X original magnification (e); f, FACS analysis of mechanism of cell death of the sst1 congenic macrophages infected with MTB (top panels) or BCG (bottom panels) in vitro; g, Multiplication of MTB (left panel) or M. bovis BCG (right panel) in the sst1 congenic macrophages in vitro (*p<0.01, **p<0.001). Error bars represent 95% confidence intervals.
Figure 2. Identification of the _sst1_candidate gene.
a, Physical map of the sst1 minimal region. (#)- number of recombination evens, (M) – polymorphic markers, (C) – chromosome with distances between the markers (kb); (G) – known genes; (RC) - recombinant chromosomes containing the sst1 resistant (R) or susceptible (S) alleles, genotypes for each marker are represented by solid (B6) and opened (C3H) boxes; b, Analysis of the _Ifi75_-rs expression in the tuberculosis lung lesions of the sst1 congenic mice by RACE. c, Domain structure of the Ipr1 and its human homolog SP110b and location of the PCR primers. d, Ipr1 and Sp100-rs gene expression in the lungs during MTB infection (Northern blot); e, Ipr1 gene expression in sst1 S (S) or sst1 R (R) macrophages infected MTB, BCG or activated with IFN-γ in vitro.
Figure 3. Lack of the Ipr1 gene expression in the C3HeB/FeJ substrain correlates with its extreme susceptibility to MTB infection.
a, Survival after the intravenous infection with MTB; b, MTB bacterial loads three weeks after the infection (4 mice per strain, ** p<0.001, error bars represent standard deviation); c, Analysis of the _sst1_-encoded candidate gene expression in the tuberculosis lung lesions by RT-PCR three weeks after the infection.
Figure 4. Expression of the Ipr1 transgene in the sst1S macrophages confers resistance to intracellular pathogens MTB and L. monocytogenes.
a, RT-PCR of Ipr1 and Sp100-rs in macrophages isolated from the sst1 S (S), sst1 R(R) mice, their F1 hybrids (SxR, RxS) and the Ipr1 transgenic (Tg) mice, 1 - IFNγ -stimulated, 2 - MTB-infected b, MTB bacterial loads in the Ipr1 transgenic (Tg+/−) and control (Tg−/−) mice after infection with MTB (7 mice/strain, * p<0.05); c, e growth of MTB (c) and L.monocytogenes (e) in the Ipr1 transgenic and control (sst1 S) macrophages (three experiments were performed in triplicates, *p<0.01, **p<0.001, error bars represent 95% confidence interval).
Comment in
- Tuberculosis: the genetics of vulnerability.
Jabado N, Gros P. Jabado N, et al. Nature. 2005 Apr 7;434(7034):709-11. doi: 10.1038/434709a. Nature. 2005. PMID: 15815612 No abstract available.
Similar articles
- Genetic dissection of host resistance to Mycobacterium tuberculosis: the sst1 locus and the Ipr1 gene.
Kramnik I. Kramnik I. Curr Top Microbiol Immunol. 2008;321:123-48. doi: 10.1007/978-3-540-75203-5_6. Curr Top Microbiol Immunol. 2008. PMID: 18727490 Review. - Tuberculosis: the genetics of vulnerability.
Jabado N, Gros P. Jabado N, et al. Nature. 2005 Apr 7;434(7034):709-11. doi: 10.1038/434709a. Nature. 2005. PMID: 15815612 No abstract available. - Characterization of promoter of the tuberculosis-resistant gene intracellular pathogen resistance 1.
Wu Y, Liu F, Zhang Y, Wang Y, Guo Z, Zhang Y. Wu Y, et al. Immunol Res. 2016 Feb;64(1):143-54. doi: 10.1007/s12026-015-8732-3. Immunol Res. 2016. PMID: 26590945 - The host resistance locus sst1 controls innate immunity to Listeria monocytogenes infection in immunodeficient mice.
Boyartchuk V, Rojas M, Yan BS, Jobe O, Hurt N, Dorfman DM, Higgins DE, Dietrich WF, Kramnik I. Boyartchuk V, et al. J Immunol. 2004 Oct 15;173(8):5112-20. doi: 10.4049/jimmunol.173.8.5112. J Immunol. 2004. PMID: 15470055 - [Protective immunity against Mycobacterium tuberculosis].
Kawamura I. Kawamura I. Kekkaku. 2006 Nov;81(11):687-91. Kekkaku. 2006. PMID: 17154048 Review. Japanese.
Cited by
- Mouse model of pulmonary cavitary tuberculosis and expression of matrix metalloproteinase-9.
Ordonez AA, Tasneen R, Pokkali S, Xu Z, Converse PJ, Klunk MH, Mollura DJ, Nuermberger EL, Jain SK. Ordonez AA, et al. Dis Model Mech. 2016 Jul 1;9(7):779-88. doi: 10.1242/dmm.025643. Epub 2016 May 26. Dis Model Mech. 2016. PMID: 27482816 Free PMC article. - Host-Directed Therapies for Tuberculosis.
Tobin DM. Tobin DM. Cold Spring Harb Perspect Med. 2015 May 18;5(10):a021196. doi: 10.1101/cshperspect.a021196. Cold Spring Harb Perspect Med. 2015. PMID: 25986592 Free PMC article. Review. - Escape from the Phagosome: The Explanation for MHC-I Processing of Mycobacterial Antigens?
Harriff MJ, Purdy GE, Lewinsohn DM. Harriff MJ, et al. Front Immunol. 2012 Mar 5;3:40. doi: 10.3389/fimmu.2012.00040. eCollection 2012. Front Immunol. 2012. PMID: 22566923 Free PMC article. - Host innate immune response to Mycobacterium tuberculosis.
Bhatt K, Salgame P. Bhatt K, et al. J Clin Immunol. 2007 Jul;27(4):347-62. doi: 10.1007/s10875-007-9084-0. Epub 2007 Mar 16. J Clin Immunol. 2007. PMID: 17364232 Review. - In mice, tuberculosis progression is associated with intensive inflammatory response and the accumulation of Gr-1 cells in the lungs.
Lyadova IV, Tsiganov EN, Kapina MA, Shepelkova GS, Sosunov VV, Radaeva TV, Majorov KB, Shmitova NS, van den Ham HJ, Ganusov VV, De Boer RJ, Racine R, Winslow GM. Lyadova IV, et al. PLoS One. 2010 May 4;5(5):e10469. doi: 10.1371/journal.pone.0010469. PLoS One. 2010. PMID: 20454613 Free PMC article.
References
- Raviglione MC. The TB epidemic from 1992 to 2002. Tuberculosis (Edinb) 2003;83:4–14. - PubMed
- Casanova JL, Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu Rev Immunol. 2002;20:581–620. - PubMed
- Bleed D, Dye C, Raviglione MC. Dynamics and control of the global tuberculosis epidemic. Curr Opin Pulm Med. 2000;6:174–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases