Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation - PubMed (original) (raw)
Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation
Balaraj B Menon et al. Proc Natl Acad Sci U S A. 2005.
Abstract
The recruitment model for gene activation presumes that DNA is a platform on which the requisite components of the transcriptional machinery are assembled. In contrast to this idea, we show here that Rap1/Gcr1/Gcr2 transcriptional activation in yeast cells occurs through a large anchored protein platform, the Nup84 nuclear pore subcomplex. Surprisingly, Nup84 and associated subcomplex components activate transcription themselves in vivo when fused to a heterologous DNA-binding domain. The Rap1 coactivators Gcr1 and Gcr2 form an important bridge between the yeast nuclear pore complex and the transcriptional machinery. Nucleoporin activation may be a widespread eukaryotic phenomenon, because it was first detected as a consequence of oncogenic rearrangements in acute myeloid leukemia and related syndromes in humans. These chromosomal translocations fuse a homeobox DNA-binding domain to the human homolog (hNup98) of a transcriptionally active component of the yeast Nup84 subcomplex. We conclude that Rap1 target genes are activated by moving to contact compartmentalized nuclear assemblages, rather than through recruitment of the requisite factors to chromatin by means of diffusion. We term this previously undescribed mechanism "reverse recruitment" and discuss the possibility that it is a central feature of eukaryotic gene regulation. Reverse recruitment stipulates that activators work by bringing the DNA to an nuclear pore complex-tethered platform of assembled transcriptional machine components.
Figures
Fig. 1.
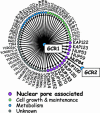
Genome-wide genetic analysis of GCR1. A Δ_gcr1_ query strain (SD8) was crossed to an array of ≈4,700 deletion mutants to screen for synthetic growth defects in the resulting double mutants. Synthetic defects that result from combination with GCR1 deletion (arrows) fall into four categories based on Gene Ontology annotation (29) as follows: nuclear pore-associated genes (purple; large type), genes involved in cell growth and maintenance (light green), metabolic genes (blue), and uncharacterized ORFs (gray). Components of the Nup84 nuclear pore subcomplex and the related factor Nup100 are shown in bold. Deletion of NUP84 is also synthetically defective in combination with a GCR2 deletion, as indicated.
Fig. 2.
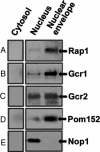
Physical association between the Rap1/Gcr1/Gcr2 activation complex and the nuclear periphery. Rap1 (A), Gcr1-myc (B), and Gcr2-myc (C) copurify with Pom152 (D) in nuclear envelope fractions; the nucleolar protein Nop1 (E) served as a negative control.
Fig. 3.
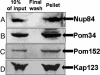
Epitope-tagged Gcr1 coimmunoprecipitates with three nuclear pore factors [Nup84 (A), Pom34 (B), and Pom152 (C)] and the NPC-associated β-importin Kap123 (D). Gcr1-myc was immunoprecipitated from whole cell extracts (input; lane 1); the last of four washes before elution (final wash; lane 2) was analyzed as a control. The corresponding protein eluates (pellet) were loaded in lane 3.
Fig. 4.
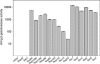
Transcriptional activation at the nuclear rim. The exclusively cytoplasmic nucleoporin Nup42 and the Seh1 component of the Nup84 subcomplex failed to activate transcription above background levels (vector) in WT cells. All other nucleoporins tested, including the human nucleoporin hNup98 (protooncogenic homolog of Nup145C), stimulated transcription of the reporter gene. The conventional activators Gcr1, Gcr2, Gcn4, and Swi4, as well as the mediator components Ssn8 and Sin4, are shown for comparison. Error bars represent standard error of the mean.
Fig. 5.
Activation and repression by the multifunctional regulator Rap1 at the nuclear periphery of S. cerevisiae. Known components of the Rap1 activation (Gcr1/Gcr2; refs. –, , , and 19) or silencing (Sir complex; refs. –, , , and 19) assemblages are shown. Essential nucleoporins are indicated with an asterisk; components of the GCR1 genetic network identified by SGA analysis (Fig. 1) are shown in bold. Note that combining deletion of GCR1 with deletion of each of several genes encoding components of the Rap1 silencing assemblage did not result in a synthetic growth defect (NUP2, NUP60, YKU70, YKU80, and SIR1 were tested by tetrad dissection). Dashed lines highlight presumptive perinuclear tethering interactions; NPC-associated factors shown to interact with Gcr1 (Fig. 3) are underlined. The representation shown here is not intended to rule out the existence of a unified complex that can switch between activation and repression of transcription (see text for further discussion).
Similar articles
- Role of the N-terminal region of Rap1p in the transcriptional activation of glycolytic genes in Saccharomyces cerevisiae.
Mizuno T, Kishimoto T, Shinzato T, Haw R, Chambers A, Wood J, Sinclair D, Uemura H. Mizuno T, et al. Yeast. 2004 Jul 30;21(10):851-66. doi: 10.1002/yea.1123. Yeast. 2004. PMID: 15300680 - Identification of a transcriptional activation domain in yeast repressor activator protein 1 (Rap1) using an altered DNA-binding specificity variant.
Johnson AN, Weil PA. Johnson AN, et al. J Biol Chem. 2017 Apr 7;292(14):5705-5723. doi: 10.1074/jbc.M117.779181. Epub 2017 Feb 14. J Biol Chem. 2017. PMID: 28196871 Free PMC article. - Role of GCR2 in transcriptional activation of yeast glycolytic genes.
Uemura H, Jigami Y. Uemura H, et al. Mol Cell Biol. 1992 Sep;12(9):3834-42. doi: 10.1128/mcb.12.9.3834-3842.1992. Mol Cell Biol. 1992. PMID: 1508187 Free PMC article. - The multifunctional transcription factor Rap1: a regulator of yeast physiology.
Azad GK, Tomar RS. Azad GK, et al. Front Biosci (Landmark Ed). 2016 Jun 1;21(5):918-30. doi: 10.2741/4429. Front Biosci (Landmark Ed). 2016. PMID: 27100480 Review. - Transcribe this way: Rap1 confers promoter directionality by repressing divergent transcription.
Wu ACK, Van Werven FJ. Wu ACK, et al. Transcription. 2019 Jun;10(3):164-170. doi: 10.1080/21541264.2019.1608716. Epub 2019 May 5. Transcription. 2019. PMID: 31057041 Free PMC article. Review.
Cited by
- Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes.
Palancade B, Liu X, Garcia-Rubio M, Aguilera A, Zhao X, Doye V. Palancade B, et al. Mol Biol Cell. 2007 Aug;18(8):2912-23. doi: 10.1091/mbc.e07-02-0123. Epub 2007 May 30. Mol Biol Cell. 2007. PMID: 17538013 Free PMC article. - NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus.
Saito K, Yoshikawa M, Yano K, Miwa H, Uchida H, Asamizu E, Sato S, Tabata S, Imaizumi-Anraku H, Umehara Y, Kouchi H, Murooka Y, Szczyglowski K, Downie JA, Parniske M, Hayashi M, Kawaguchi M. Saito K, et al. Plant Cell. 2007 Feb;19(2):610-24. doi: 10.1105/tpc.106.046938. Epub 2007 Feb 16. Plant Cell. 2007. PMID: 17307929 Free PMC article. - Sus1, Sac3, and Thp1 mediate post-transcriptional tethering of active genes to the nuclear rim as well as to non-nascent mRNP.
Chekanova JA, Abruzzi KC, Rosbash M, Belostotsky DA. Chekanova JA, et al. RNA. 2008 Jan;14(1):66-77. doi: 10.1261/rna.764108. Epub 2007 Nov 14. RNA. 2008. PMID: 18003937 Free PMC article. - Nuclear pore interactions with the genome.
Sood V, Brickner JH. Sood V, et al. Curr Opin Genet Dev. 2014 Apr;25:43-9. doi: 10.1016/j.gde.2013.11.018. Epub 2014 Jan 28. Curr Opin Genet Dev. 2014. PMID: 24480294 Free PMC article. Review. - A nuclear pore sub-complex restricts the propagation of Ty retrotransposons by limiting their transcription.
Bonnet A, Chaput C, Palmic N, Palancade B, Lesage P. Bonnet A, et al. PLoS Genet. 2021 Nov 1;17(11):e1009889. doi: 10.1371/journal.pgen.1009889. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34723966 Free PMC article.
References
- Cosma, M. P. (2002) Mol. Cell 10, 227–236. - PubMed
- Bryant, G. O. & Ptashne, M. (2003) Mol. Cell 11, 1301–1309. - PubMed
- Ptashne, M. (2003) Philos. Trans. R. Soc. London 361, 1223–1234. - PubMed
- Ptashne, M. & Gann, A. (1997) Nature 386, 569–577. - PubMed
- Struhl, K., Kadosh, D., Keaveney, M., Kuras, L. & Moqtaderi, Z. (1998) Cold Spring Harbor Symp. Quant. Biol. 63, 413–421. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials