Deduction of probable events of lateral gene transfer through comparison of phylogenetic trees by recursive consolidation and rearrangement - PubMed (original) (raw)
Deduction of probable events of lateral gene transfer through comparison of phylogenetic trees by recursive consolidation and rearrangement
Dave MacLeod et al. BMC Evol Biol. 2005.
Abstract
Background: When organismal phylogenies based on sequences of single marker genes are poorly resolved, a logical approach is to add more markers, on the assumption that weak but congruent phylogenetic signal will be reinforced in such multigene trees. Such approaches are valid only when the several markers indeed have identical phylogenies, an issue which many multigene methods (such as the use of concatenated gene sequences or the assembly of supertrees) do not directly address. Indeed, even when the true history is a mixture of vertical descent for some genes and lateral gene transfer (LGT) for others, such methods produce unique topologies.
Results: We have developed software that aims to extract evidence for vertical and lateral inheritance from a set of gene trees compared against an arbitrary reference tree. This evidence is then displayed as a synthesis showing support over the tree for vertical inheritance, overlaid with explicit lateral gene transfer (LGT) events inferred to have occurred over the history of the tree. Like splits-tree methods, one can thus identify nodes at which conflict occurs. Additionally one can make reasonable inferences about vertical and lateral signal, assigning putative donors and recipients.
Conclusion: A tool such as ours can serve to explore the reticulated dimensionality of molecular evolution, by dissecting vertical and lateral inheritance at high resolution. By this, we mean that individual nodes can be examined not only for congruence, but also for coherence in light of LGT. We assert that our tools will facilitate the comparison of phylogenetic trees, and the interpretation of conflicting data.
Figures
Figure 2
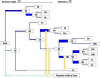
Synthesis diagram. The vertical-inheritance backbone representing the input reference tree is shown in dark blue, with the line thickness of an internal branch corresponding to the frequency of its support across the whole dataset. Putative LGT events are in orange, connecting donors (circles) with recipients (arrowheads); where there are multiple possible donor candidates, these converge onto a double arrowhead (see text). Where the apparent donor of a gene falls outside of the taxa included in the analysis, one is created as a basal group taxon, indicated in light blue. In order to avoid graphical congestion, branches in the tree may be artificially extended, as dotted segments. Colours are editable, and links are interactive. Clicking on node 3A, for example, displays the following message: 3A: set006r, branch length:0.03755, thickness:0.25, files supported: mvin.tre:2/2, where support for the node is 2/2 for mvin.tre (both edit paths support the node), and 0 (unsupported) for hypoprot.tre, biob.tre and n6methylase.tre. The segment's thickness, therefore, is simply (2/2)/4.
Figure 1
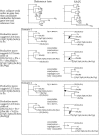
Step-by-step example of the consolidation and rearrangement method of Horizstory. Where indicated as resolved, nodes were supported by a bootstrap value better than 80%. Two minimal scenarios were found in the analysis of the MviN tree shown in this example, both suggesting an equivalent pair of lateral gene transfer events (as illustrated in Lumbermill, Figure 2). Other (longer or unproductive) scenarios are not shown. A productive move is one where the gene tree gains further similarity to the reference tree, permitting further consolidation (see text). Ba, Buchnera aphidicola; Ec, Escherichia coli; Hi, Haemophilus influenzae; Pa, Pseudomonas aeruginosa; Pm, Pasteurella multocida; Se, Salmonella enterica; Vc, Vibrio cholerae; Wg, Wigglesworthia glossinidia; Xa, Xanthomonas axonopodis; Xc, Xanthomonas campestris; Xf, Xylella fastidiosa; YpC, Yersinia pestis CO92; YpK, Yersinia pestis KIM.
Figure 3
Estimate of genomic mosaicism. This application screenshot displays a table indicating the degree of vertical and of horizontal inheritance inferred for each node in the synthesis (see Fig. 2). The middle two columns of the table summarize the distribution of vertical (blue) and of lateral (orange) support for the segment immediately preceding each node, and the vertical and lateral support for the whole string of segments leading to a particular node, respectively. For example, B. aphidicola shows a 14% degree of accumulated mosaicism since the tree's root (from the small sample of genes used in this analysis), but no recent mosaicism (in the segment preceding its divergence from W. glossinidia, node 5A). Numerical values for the proportions are given by clicking on a bar. Node 1B is left entirely blank for want of phylogenetic evidence.
Similar articles
- An emerging phylogenetic core of Archaea: phylogenies of transcription and translation machineries converge following addition of new genome sequences.
Brochier C, Forterre P, Gribaldo S. Brochier C, et al. BMC Evol Biol. 2005 Jun 2;5:36. doi: 10.1186/1471-2148-5-36. BMC Evol Biol. 2005. PMID: 15932645 Free PMC article. - Detecting lateral genetic transfer : a phylogenetic approach.
Beiko RG, Ragan MA. Beiko RG, et al. Methods Mol Biol. 2008;452:457-69. doi: 10.1007/978-1-60327-159-2_21. Methods Mol Biol. 2008. PMID: 18566777 Review. - Scaling Up the Phylogenetic Detection of Lateral Gene Transfer Events.
Chan CX, Beiko RG, Ragan MA. Chan CX, et al. Methods Mol Biol. 2017;1525:421-432. doi: 10.1007/978-1-4939-6622-6_16. Methods Mol Biol. 2017. PMID: 27896730 - Phylogenomic networks.
Dagan T. Dagan T. Trends Microbiol. 2011 Oct;19(10):483-91. doi: 10.1016/j.tim.2011.07.001. Epub 2011 Aug 3. Trends Microbiol. 2011. PMID: 21820313 Review.
Cited by
- Bioinformatic detection of horizontally transferred DNA in bacterial genomes.
Langille MG, Brinkman FS. Langille MG, et al. F1000 Biol Rep. 2009 Mar 24;1:25. doi: 10.3410/B1-25. F1000 Biol Rep. 2009. PMID: 20948661 Free PMC article. - Inferring horizontal gene transfer.
Ravenhall M, Škunca N, Lassalle F, Dessimoz C. Ravenhall M, et al. PLoS Comput Biol. 2015 May 28;11(5):e1004095. doi: 10.1371/journal.pcbi.1004095. eCollection 2015 May. PLoS Comput Biol. 2015. PMID: 26020646 Free PMC article. - MimiLook: A Phylogenetic Workflow for Detection of Gene Acquisition in Major Orthologous Groups of Megavirales.
Jain S, Panda A, Colson P, Raoult D, Pontarotti P. Jain S, et al. Viruses. 2017 Apr 7;9(4):72. doi: 10.3390/v9040072. Viruses. 2017. PMID: 28387730 Free PMC article. - Parsimonious inference of hybridization in the presence of incomplete lineage sorting.
Yu Y, Barnett RM, Nakhleh L. Yu Y, et al. Syst Biol. 2013 Sep;62(5):738-51. doi: 10.1093/sysbio/syt037. Epub 2013 Jun 4. Syst Biol. 2013. PMID: 23736104 Free PMC article. - Do orthologous gene phylogenies really support tree-thinking?
Bapteste E, Susko E, Leigh J, MacLeod D, Charlebois RL, Doolittle WF. Bapteste E, et al. BMC Evol Biol. 2005 May 24;5:33. doi: 10.1186/1471-2148-5-33. BMC Evol Biol. 2005. PMID: 15913459 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases