Proteomic and genomic characterization of chromatin complexes at a boundary - PubMed (original) (raw)
Proteomic and genomic characterization of chromatin complexes at a boundary
Alan J Tackett et al. J Cell Biol. 2005.
Abstract
We have dissected specialized assemblies on the Saccharomyces cerevisiae genome that help define and preserve the boundaries that separate silent and active chromatin. These assemblies contain characteristic stretches of DNA that flank particular regions of silent chromatin, as well as five distinctively modified histones and a set of protein complexes. The complexes consist of at least 15 chromatin-associated proteins, including DNA pol epsilon, the Isw2-Itc1 and Top2 chromatin remodeling proteins, the Sas3-Spt16 chromatin modifying complex, and Yta7, a bromodomain-containing AAA ATPase. We show that these complexes are important for the faithful maintenance of an established boundary, as disruption of the complexes results in specific, anomalous alterations of the silent and active epigenetic states.
Figures
Figure 1.
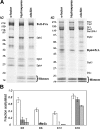
Dpb4 forms multiple stable complexes with chromatin. (A) Complexes were isolated via a PrA tag under conditions that copurified interacting proteins. These proteins were resolved by SDS-PAGE, visualized by Coomassie blue staining, and excised for MS identification. Labels show specific interactions with the PrA-tagged proteins. Reciprocal purifications of tagged members of the complexes demonstrated coisolation of Dpb4 with the exception of tagged Yta7. (B) Coimmunoprecipitation experiments with Yta7 and Dpb4 indicated that they associate in vivo. Note lane 7, which controls for post-lysis association of Yta7 and Dpb4 in vitro.
Figure 2.
Elucidating the architecture of chromatin-associated complexes. (A) Dpb3 or Dpb4 was separately deleted in combination with the various PrA-tagged pol ɛ components. PrA-tagged components and their interacting proteins were isolated and visualized as for Fig. 1 A. Gel positions shown with asterisks, corresponding to the migration position of the proteins identified in Fig. 1, were excised and screened for the presence of these proteins by MS. (B) Proposed assembly pathway for chromatin complexes containing Dpb4.
Figure 3.
Isolation of chromatin-associated protein complexes with their cognate DNA and histones. Complexes containing Dpb4-PrA were immunoisolated and eluted under nondenaturing conditions. (A) The eluate was loaded on an anion exchange column and fractions were collected from a 0–1 M [NaCl] gradient and visualized by Coomassie blue–stained SDS-PAGE. DNA polymerase activity was assayed by dTMP incorporation into calf thymus DNA. The presence of copurifying DNA was assayed by real-time PCR. Lanes containing three unique complexes are shown expanded, and the protein components identified by MS are labeled. Bands labeled with an asterisk may represent a breakdown product or alternate form of the protein. White lines indicate that intervening lanes have been spliced out. (B) The three unique complexes from A were further resolved by gel filtration. Fraction proteins were visualized by silver-stained SDS-PAGE.
Figure 4.
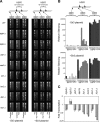
Pol ɛ and the chromatin-remodeling complexes are associated with a specific epigenetic state of chromatin. Histones from the Dpb4-chromatin remodeling complex and from the Dpb4–pol ɛ complex were PrA-affinity purified from _DPB4-PrA dpb3_Δ and DPB3-PrA strains, respectively. Histone H4 was modified in-gel with D6-acetic anhydride to convert free lysines to D3-acetyl lysines and then digested with trypsin. (A) MALDI-QqTOF mass spectrum of the tryptic peptide encompassing amino acid residues 4–17 of the NH2-terminal tail of histone H4. All five possible acetylation states are labeled. (B) Each peptide ion species observed in A was fragmented in a MALDI-ion trap MS. The resulting heavy and light fragment ion intensities were used to determine the levels of acetylation on specific lysines. Shown is a representative fragmentation spectrum of peptide 4–17 containing two heavy (Acd) and two light (Ac) acetylations. The b- and y-fragment ions used to quantitate the levels of acetylation on each of the four lysines are shown expanded. (C) Site-specific levels of acetylation on histone H4 peptide 4–17 from global histones (gray), Dpb4-chromatin remodeling complex-associated histones (light blue), pol ɛ–associated histones from Dpb3-PrA immunoprecipitation (medium blue), and pol ɛ–associated histones from Pol2-PrA immunoprecipitation (dark blue). (D) Site-specific levels of acetylation on histone H4 peptide 4–17 from immunoprecipitation of Pol2-PrA (dark blue) and Pol2-PrA in _dpb3_Δ strain (green).
Figure 5.
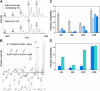
Genome-wide localization of Dpb4-containing complexes. Protected DNA from the purified fraction of Dpb4-associated histones in Fig. 3 A was hybridized to an intergenic DNA microarray covering the S. cerevisiae genome. (A) The sequences of chromosomes I–XVI are depicted as horizontal black lines with the centromeres denoted by circles. Vertical ticks indicate the position of sequences enriched with the Dpb4-histone complexes. The positions of two silenced gene clusters (HM and FLO) are indicated, showing their close apposition to the Dpb4-enriched sequences. (B) A histogram showing the number of intergenic regions enriched with the Dpb4-histone complexes is plotted as a function of distance from the chromosome ends. (C) Dpb4-histone complex DNA is enriched at boundaries to the silent, mating loci on chromosome III. Positions of enriched sequences are shown as horizontal black lines above the annotated chromosome with increasing line thickness indicating increasing enrichment. The inset shows high resolution mapping of Dpb4-enriched sequences from semiquantitative PCR analysis. The asterisk marks a region that was not covered by the microarray analysis.
Figure 6.
Dpb4-containing chromatin complexes regulate an HMR boundary. (A) Strains carrying deletions of DPB4, DPB3, YTA7, SAS3, ITC1, ISW2, DLS1, and HTZ1 were assayed for the expression of a URA3 reporter gene placed in the following three locations: in the Sir-silenced region of HMR (∼640 bp to the right of HMR-E), at the left-hand boundary (∼475 bp to the left of HMR-E), and upstream of the boundary region (∼2,840 bp to the left of HMR-E, and within the YCR095C gene) (Donze et al., 1999). Decreased silencing of the reporter (i.e., increased transcription) results in increased cell death on FOA. Strains were assayed either without or with a Sir3-expressing plasmid. (B) Semiquantitative relative measure of the dilution-adjusted colony density seen in A. (C) Transcription levels of YCR095C (light gray) and GIT1 (dark gray), genes proximal to HMR, were measured by real-time PCR. Fold transcription is relative to wild type. Measurements less than onefold indicate repression of transcription, whereas measurements greater than onefold indicate above normal transcription. Error bars show the SD of the mean for triplicate measurements.
Figure 7.
Pol ɛ associates with chromatin during DNA replication. (A) Pol2-PrA or Dpb4-PrA complexes were affinity purified at different cell cycle blocks, and their proteins were resolved by Coomassie blue–stained SDS-PAGE. White lines indicate that intervening lanes have been spliced out. (B) Site-specific levels of acetylation on histone H4 peptide 4–17 from the Pol2-PrA–associated histones from global (open), hydroxyurea block (dark gray), and _cdc20_Δ block (light gray).
Figure 8.
A model for the roles of the Dpb4-associated complexes at a chromatin boundary. The mating genes at HMR are contained within a region flanked by Orc complexes and transcriptionally silenced through the binding of Sir proteins. The Dpb4-containing pol ɛ and chromatin modifying/remodeling complexes associate with boundary regions where they bind distinctively modified histones. The pol ɛ complex aids in duplication of the silent chromatin, whereas the Dpb4-chromatin remodeling complex preserves the boundaries. Precise protein positions and their oligomeric states have not been determined.
Similar articles
- Genome-wide patterns of histone modifications in yeast.
Millar CB, Grunstein M. Millar CB, et al. Nat Rev Mol Cell Biol. 2006 Sep;7(9):657-66. doi: 10.1038/nrm1986. Epub 2006 Aug 16. Nat Rev Mol Cell Biol. 2006. PMID: 16912715 Review. - A noncanonical bromodomain in the AAA ATPase protein Yta7 directs chromosomal positioning and barrier chromatin activity.
Gradolatto A, Smart SK, Byrum S, Blair LP, Rogers RS, Kolar EA, Lavender H, Larson SK, Aitchison JD, Taverna SD, Tackett AJ. Gradolatto A, et al. Mol Cell Biol. 2009 Sep;29(17):4604-11. doi: 10.1128/MCB.00160-09. Epub 2009 Jul 6. Mol Cell Biol. 2009. PMID: 19581291 Free PMC article. - The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae.
Meijsing SH, Ehrenhofer-Murray AE. Meijsing SH, et al. Genes Dev. 2001 Dec 1;15(23):3169-82. doi: 10.1101/gad.929001. Genes Dev. 2001. PMID: 11731480 Free PMC article. - Using genomics and proteomics to investigate mechanisms of transcriptional silencing in Saccharomyces cerevisiae.
Gao L, Gross DS. Gao L, et al. Brief Funct Genomic Proteomic. 2006 Dec;5(4):280-8. doi: 10.1093/bfgp/ell035. Epub 2006 Nov 2. Brief Funct Genomic Proteomic. 2006. PMID: 17082210 Review. - Saccharomyces cerevisiae Yta7 regulates histone gene expression.
Gradolatto A, Rogers RS, Lavender H, Taverna SD, Allis CD, Aitchison JD, Tackett AJ. Gradolatto A, et al. Genetics. 2008 May;179(1):291-304. doi: 10.1534/genetics.107.086520. Genetics. 2008. PMID: 18493054 Free PMC article.
Cited by
- Unraveling the histone's potential: a proteomics perspective.
Brumbaugh J, Phanstiel D, Coon JJ. Brumbaugh J, et al. Epigenetics. 2008 Sep;3(5):254-7. doi: 10.4161/epi.3.5.7005. Epub 2008 Sep 17. Epigenetics. 2008. PMID: 18849650 Free PMC article. - Quantitative cell array screening to identify regulators of gene expression.
Kainth P, Andrews B. Kainth P, et al. Brief Funct Genomics. 2010 Jan;9(1):13-23. doi: 10.1093/bfgp/elp047. Epub 2009 Dec 1. Brief Funct Genomics. 2010. PMID: 19952074 Free PMC article. Review. - CATP is a critical component of the Neurospora circadian clock by regulating the nucleosome occupancy rhythm at the frequency locus.
Cha J, Zhou M, Liu Y. Cha J, et al. EMBO Rep. 2013 Oct;14(10):923-30. doi: 10.1038/embor.2013.131. Epub 2013 Aug 20. EMBO Rep. 2013. PMID: 23958634 Free PMC article. - Comprehensive analysis of interacting proteins and genome-wide location studies of the Sas3-dependent NuA3 histone acetyltransferase complex.
Vicente-Muñoz S, Romero P, Magraner-Pardo L, Martinez-Jimenez CP, Tordera V, Pamblanco M. Vicente-Muñoz S, et al. FEBS Open Bio. 2014 Nov 8;4:996-1006. doi: 10.1016/j.fob.2014.11.001. eCollection 2014. FEBS Open Bio. 2014. PMID: 25473596 Free PMC article. - ChAP-MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus.
Byrum SD, Raman A, Taverna SD, Tackett AJ. Byrum SD, et al. Cell Rep. 2012 Jul 26;2(1):198-205. doi: 10.1016/j.celrep.2012.06.019. Epub 2012 Jul 20. Cell Rep. 2012. PMID: 22840409 Free PMC article.
References
- Aitchison, J.D., M.P. Rout, M. Marelli, G. Blobel, and R.W. Wozniak. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131:1133–1148. - PMC - PubMed
- Aitchison, J.D., G. Blobel, and M.P. Rout. 1996. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 274:624–627. - PubMed
- Archambault, V., E.J. Chang, B.J. Drapkin, F.R. Cross, B.T. Chait, and M.P. Rout. 2004. Targeted proteomic study of the cyclin-cdk module. Mol. Cell. 14:699–711. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM038839/GM/NIGMS NIH HHS/United States
- R33 CA089810/CA/NCI NIH HHS/United States
- P41 RR000862/RR/NCRR NIH HHS/United States
- CA89810/CA/NCI NIH HHS/United States
- GM38839/GM/NIGMS NIH HHS/United States
- RR00862/RR/NCRR NIH HHS/United States
- R01 GM038839/GM/NIGMS NIH HHS/United States
- GM066496/GM/NIGMS NIH HHS/United States
- F32 GM066496/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous