An internal GAP domain negatively regulates presynaptic dynamin in vivo: a two-step model for dynamin function - PubMed (original) (raw)
An internal GAP domain negatively regulates presynaptic dynamin in vivo: a two-step model for dynamin function
Radhakrishnan Narayanan et al. J Cell Biol. 2005.
Abstract
The mechanism by which the self-assembling GTPase dynamin functions in vesicle formation remains controversial. Point mutations in shibire, the Drosophila dynamin, cause temperature-sensitive (ts) defects in endocytosis. We show that the ts2 mutation, which occurs in the switch 2 region of dynamin's GTPase domain, compromises GTP binding affinity. Three second-site suppressor mutations, one in the switch 1 region of the GTPase domain and two in the GTPase effector domain (GED), dynamin's putative GAP, fully rescue the shi(ts2) defects in synaptic vesicle recycling. The functional rescue in vivo correlates with a reduction in both the basal and assembly-stimulated GTPase activity in vitro. These findings demonstrate that GED is indeed an internal dynamin GAP and establish that, as for other GTPase superfamily members, dynamin's function in vivo is negatively regulated by its GAP activity. Based on these and other observations, we propose a two-step model for dynamin during vesicle formation in which an early regulatory GTPase-like function precedes late, assembly-dependent steps during which GTP hydrolysis is required for vesicle release.
Figures
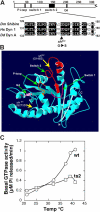
Figure 1.
The shits2 mutation (G146S) is in the highly conserved switch 2 region of the GTPase domain. (A) Alignment of Drosophila shibire, human dynamin-1, and Dictyostelium dynamin A around the ts2 mutation shows tightly conserved residues. (B) Threaded structure of human dynamin-1 GTPase domain folded on a template crystal structure of Dictyostelium dynamin A GTPase domain (Niemann et al., 2001) using Geno3D automatic comparative modeling of three-dimensional protein structure (Combet et al., 2002) and Swiss-PdbViewer (
). The four consensus GTP binding elements (green arrows) and the locations of the ts1 and ts2 mutations (yellow arrows) are indicated. (C) Temperature dependence of basal GTPase activity of dyn1:wt (○) and dyn1:ts2 (□) measured in the presence of 100 μM GTP.
Figure 2.
Dyn1:ts2 has a ts defect in GTP binding. (A) The basal rates of GTP hydrolysis were determined in the presence of varying concentrations of GTP for 0.5 μM dyn1:wt (○, •) or dyn1:ts2 (□, ▪) at 20 and 39°C, as described in Materials and methods_._ Shown are averaged data from three independent experiments ± SEM. The kcat (B) and Km (C) values for basal GTPase activity were calculated from the data in A. Errors are SDs from the best-fit curve to the data. (D) Time course of GTP hydrolysis at 39°C for dyn1:wt (•) and dyn1:ts2 (▪) measured in the presence of physiological concentrations of GTP (100 μM). Shown are averaged data from three independent experiments ± SEM. (E–H) Same as for A–D, except that assembly-stimulated GTPase activity was measured in the presence of 0.1 mg/ml lipid nanotubules and 0.1 μM dynamin.
Figure 3.
Dyn1:ts2 and the Sushi mutants exhibit wt self-assembly activities. (A) Wt and mutant dynamin proteins (2 μM) were incubated in 20 mM Hepes, pH 7.5, 2 mM MgCl2, and 10 mM KCl for 20 min at 39°C. Mixtures were sedimented at 14,000 rpm for 10 min at 4°C; pellets (P) and supernatants (S) were collected and subjected to SDS-PAGE. Proteins were detected by Coomassie blue staining. (B) Wt and mutant dynamin proteins were incubated as above except in buffer containing 150 mM KCl and with LTs. Pellets and supernatants after sedimentation are shown. (C) Negative-stained electron micrographs of wt and mutant dynamins assembled onto PI-4,5-P2–containing LTs. Bar, 50 nm.
Figure 4.
Sushi mutations completely suppress shits2 mutant phenotypes. (A) The three Sushi (suppressor of shits2) mutations (VS2, KVS, and Shy) do not alter levels of dynamin (top), but all completely rescue the ts paralytic behavior of shits2 mutants (bottom). (B) Synaptic vesicle endocytosis, indicated by uptake of the fluorescent endocytic tracer FM1-43 into stimulated nerve terminals, is completely blocked in shits2 but occurs efficiently in the presence of the Sushi mutations. (C) The synaptic vesicle protein synaptotagmin is trapped on the presynaptic plasma membrane and diffuses along the axon of shits2 nerve terminals stimulated at elevated temperatures (third panel), but shows normal (left two panels) synaptic vesicle distribution in the presence of the Shy, KVS, or VS2 mutations (right panels). (D) In the presence of the suppressor mutations, enhanced synaptic depression caused by use-dependent depletion of synaptic vesicles at shits2 nerve terminals stimulated at 10 Hz at 30°C is not observed (n = 5 for ts2 and VS2 and 4 for Shy and KVS). (E) Domain structure of dynamin including GTPase and GED, to which Sushi mutations VS2, KVS, and Shy are mapped. All three mutations identify residues conserved between Drosophila shibire and human dynamin-1.
Figure 5.
Second-site Sushi mutations alter basal and assembly-stimulated rates of GTP hydrolysis. (A) Unstimulated rates of GTP hydrolysis were determined at 39°C as described in Fig. 2 for dyn1:ts2/A738T_KVS_, dyn1:ts2/T749I_Shy_, and dyn1:ts2/R59C_VS2_as indicated. Data for wt and ts2 are redrawn from Fig. 2. Values shown are averages from independent experiments ± SEM, n = 3 for dyn1:ts2/T749I_Shy_ and dyn1:ts2/R59C_VS2_, n = 6 for dyn1:ts2/A738T_KVS_ because this mutant exhibited more variable results probably due to some aggregation, which would artificially enhance basal GTPase activity. The kcat (B) and Km (C) values for basal GTPase activity were calculated from the data in A. Errors are SDs from the best-fit curve to the data. (D–F) Same is in A–C except that the LT-stimulated rates of GTP hydrolysis were determined for each of the Sushi mutants in three independent experiments. (G) Filter assay for binding of [35S]GTPγS to wt and mutant dynamins at 39°C, performed as described in Materials and methods. Shown are dot blots from a single experiment performed in triplicate, with or without 2 μM dynamin and 25 μM GTPγ35S along with quantitation of the average ± SD from three such independent experiments.
Figure 6.
Distinct early and late functions for dynamin: a two-step model for dynamin function in endocytosis. A model that incorporates current and previously published observations (Kosaka and Ikeda, 1983; Hinshaw and Schmid, 1995; Takei et al., 1995; Sweitzer and Hinshaw, 1998; Sever et al., 1999; Marks et al., 2001) on dynamin's role in endocytosis. The model postulates an initial regulatory GTPase function for dynamin during a rate-determining precollar step in which the steady-state levels of dynamin GTP hydrolysis and/or dynamin•GTP are critical. These parameters are positively regulated by nucleoside diphosphate kinase (NDK) and negatively regulated by dynamin's GED, an internal GAP domain. The shits2 mutation impairs GTP binding, and the defect is restored by the three intragenic Sushi mutations, which impair GTPase activity and may function to stabilize dynamin•GTP. A subsequent post-collar step in endocytosis requires dynamin self-assembly and GTP hydrolysis, both of which are mediated, in part, by GED.
Similar articles
- Dynamin GTPase domain mutants block endocytic vesicle formation at morphologically distinct stages.
Damke H, Binns DD, Ueda H, Schmid SL, Baba T. Damke H, et al. Mol Biol Cell. 2001 Sep;12(9):2578-89. doi: 10.1091/mbc.12.9.2578. Mol Biol Cell. 2001. PMID: 11553700 Free PMC article. - GTPase activity of dynamin and resulting conformation change are essential for endocytosis.
Marks B, Stowell MH, Vallis Y, Mills IG, Gibson A, Hopkins CR, McMahon HT. Marks B, et al. Nature. 2001 Mar 8;410(6825):231-5. doi: 10.1038/35065645. Nature. 2001. PMID: 11242086 - Domain structure and intramolecular regulation of dynamin GTPase.
Muhlberg AB, Warnock DE, Schmid SL. Muhlberg AB, et al. EMBO J. 1997 Nov 17;16(22):6676-83. doi: 10.1093/emboj/16.22.6676. EMBO J. 1997. PMID: 9362482 Free PMC article. - Dynamin and its role in membrane fission.
Hinshaw JE. Hinshaw JE. Annu Rev Cell Dev Biol. 2000;16:483-519. doi: 10.1146/annurev.cellbio.16.1.483. Annu Rev Cell Dev Biol. 2000. PMID: 11031245 Free PMC article. Review. - Dynamin, endocytosis and intracellular signalling (review).
McClure SJ, Robinson PJ. McClure SJ, et al. Mol Membr Biol. 1996 Oct-Dec;13(4):189-215. doi: 10.3109/09687689609160598. Mol Membr Biol. 1996. PMID: 9116759 Review.
Cited by
- Membrane insertion of the pleckstrin homology domain variable loop 1 is critical for dynamin-catalyzed vesicle scission.
Ramachandran R, Pucadyil TJ, Liu YW, Acharya S, Leonard M, Lukiyanchuk V, Schmid SL. Ramachandran R, et al. Mol Biol Cell. 2009 Nov;20(22):4630-9. doi: 10.1091/mbc.e09-08-0683. Epub 2009 Sep 23. Mol Biol Cell. 2009. PMID: 19776347 Free PMC article. - Quantifying the dynamic interactions between a clathrin-coated pit and cargo molecules.
Weigel AV, Tamkun MM, Krapf D. Weigel AV, et al. Proc Natl Acad Sci U S A. 2013 Nov 26;110(48):E4591-600. doi: 10.1073/pnas.1315202110. Epub 2013 Nov 11. Proc Natl Acad Sci U S A. 2013. PMID: 24218552 Free PMC article. - Cargo and dynamin regulate clathrin-coated pit maturation.
Loerke D, Mettlen M, Yarar D, Jaqaman K, Jaqaman H, Danuser G, Schmid SL. Loerke D, et al. PLoS Biol. 2009 Mar 17;7(3):e57. doi: 10.1371/journal.pbio.1000057. PLoS Biol. 2009. PMID: 19296720 Free PMC article. - Dissecting dynamin's role in clathrin-mediated endocytosis.
Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. Mettlen M, et al. Biochem Soc Trans. 2009 Oct;37(Pt 5):1022-6. doi: 10.1042/BST0371022. Biochem Soc Trans. 2009. PMID: 19754444 Free PMC article. Review. - Real-time detection reveals that effectors couple dynamin's GTP-dependent conformational changes to the membrane.
Ramachandran R, Schmid SL. Ramachandran R, et al. EMBO J. 2008 Jan 9;27(1):27-37. doi: 10.1038/sj.emboj.7601961. Epub 2007 Dec 13. EMBO J. 2008. PMID: 18079695 Free PMC article.
References
- Betz, W.J., and G.S. Bewick. 1992. Optical analysis of synaptic vesicle recycling at the frog neuromuscular junction. Science. 255:200–203. - PubMed
- Binns, D.D., B. Barylko, N. Grichine, A.L. Adkinson, M.K. Helms, D.M. Jameson, J.F. Eccleston, and J.P. Albanesi. 1999. Correlation between self-association modes and GTPase activation of dynamin. J. Protein Chem. 18:277–290. - PubMed
- Carr, J.F., and J.E. Hinshaw. 1997. Dynamin assembles into spirals under physiological salt conditions upon the addition of GDP and gamma-phosphate analogues. J. Biol. Chem. 272:28030–28035. - PubMed
- Chen, M.L., D. Green, L. Liu, Y.C. Lam, L. Mukai, S. Rao, S. Ramagiri, K.S. Krishnan, J.E. Engel, J.J. Lin, and C.F. Wu. 2002. Unique biochemical and behavioral alterations in Drosophila shibirets1 mutants imply a conformational state affecting dynamin subcellular distribution and synaptic vesicle cycling. J. Neurobiol. 53:319–329. - PubMed
- Chen, M.S., R.A. Ober, C.C. Schroeder, T.W. Austin, C.A. Poodry, S.C. Wadsworth, and R.B. Vallee. 1991. Multiple forms of dynamin are encoded by shibire, a Drosophila gene involved in endocytosis. Nature. 351:583–586. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 MH061345/MH/NIMH NIH HHS/United States
- R37-MH61345/MH/NIMH NIH HHS/United States
- NS02001/NS/NINDS NIH HHS/United States
- GM42455/GM/NIGMS NIH HHS/United States
- R01 NS034889/NS/NINDS NIH HHS/United States
- R01 GM042455/GM/NIGMS NIH HHS/United States
- NS34889/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous