FAT10, a ubiquitin-independent signal for proteasomal degradation - PubMed (original) (raw)
FAT10, a ubiquitin-independent signal for proteasomal degradation
Mark Steffen Hipp et al. Mol Cell Biol. 2005 May.
Abstract
FAT10 is a small ubiquitin-like modifier that is encoded in the major histocompatibility complex and is synergistically inducible by tumor necrosis factor alpha and gamma interferon. It is composed of two ubiquitin-like domains and possesses a free C-terminal diglycine motif that is required for the formation of FAT10 conjugates. Here we show that unconjugated FAT10 and a FAT10 conjugate were rapidly degraded by the proteasome at a similar rate. Fusion of FAT10 to the N terminus of very long-lived proteins enhanced their degradation rate as potently as fusion with ubiquitin did. FAT10-green fluorescent protein fusion proteins were not cleaved but entirely degraded, suggesting that FAT10-specific deconjugating enzymes were not present in the analyzed cell lines. Interestingly, the prevention of ubiquitylation of FAT10 by mutation of all lysines or by expression in ubiquitylation-deficient cells did not affect FAT10 degradation. Thus, conjugation with FAT10 is an alternative and ubiquitin-independent targeting mechanism for degradation by the proteasome, which, in contrast to polyubiquitylation, is cytokine inducible and irreversible.
Figures
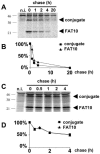
FIG. 1.
FAT10 and a FAT10 conjugate are rapidly degraded in a proteasome-dependent manner. The tetracycline-inducible FAT10 transfectant TB1N was labeled with [35S]Met-Cys and chased for the indicated time periods in the absence (A) or presence (C) of the proteasome inhibitor lactacystin (80 μM) followed by immunoprecipitation against HA-FAT10. Lanes 1 in panels A and C represent noninduced TB1N cells (n.i.). The bands shown in panels A and C were quantified on a radioimager and plotted in panels B and D, respectively, as percent radioactivity based on values obtained after the pulse.
FIG. 2.
FAT10 and ubiquitin are equally potent in targeting for degradation. (A) FAT10, SUMO, and ubiquitin were transiently expressed as uncleavable AV/GG mutated and HA-tagged GFP fusion proteins in HeLa cells. The cells were labeled for 1 h with [35S]Cys-Met and chased for the indicated time periods prior to HA-specific immunoprecipitation, SDS-polyacrylamide gel electrophoresis (PAGE), autoradiography, and quantification on a radioimager. As a control, GFP was expressed and immunoprecipitated with an anti-GFP antibody. The arrowheads denote the different GFP fusion proteins and GFP as indicated above; an asterisk indicates an unspecific band. (B) Fusion proteins of DHFR with HA-tagged FAT10 and ubiquitin as well as a DHFR-HA-UbK48R control were expressed in HeLa cells, and a pulse-chase experiment was performed as shown for panel A. The arrowheads denote the respective DHFR fusion proteins as indicated above the panels. (C) HA-FAT10-GFP and HA-ubiquitin-GFP were transiently expressed in HeLa cells without or together with HA-NUB1L. A pulse-chase analysis was performed as shown for panels A and B. An arrow denotes HA-Ub-AV-GFP, and an arrowhead denotes HA-FAT10-AV-GFP. All bands were quantified on a radioimager and plotted below each lane as percent radioactivity based on values obtained after the pulse. The data represent means of results from three independent experiments ± standard errors of the means.
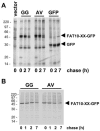
FIG. 3.
Lack of evidence for a FAT10-specific processing protease. GFP and FAT10-GFP fusion proteins with either wild-type FAT10 or with a GG-to-AV mutation of the FAT10 C terminus were transiently expressed in (A) HeLa or (B) HEK293T cells. The cells were labeled for 1 h with [35S]Cys-Met and chased for the indicated time periods prior to GFP-specific immunoprecipitation, SDS/PAGE, and autoradiography. The leftmost lane in panel A shows a vector control.
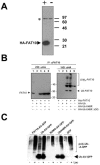
FIG. 4.
Ubiquitylation status of FAT10. (A) Characterization of a polyclonal anti-human FAT10 antibody. HEK293T cells were transiently transfected with an HA-FAT10 expression construct (+) or a vector control (−). A Western blot with a polyclonal antibody raised in rabbits against a GST-FAT10 fusion protein was then performed. The HA-FAT10 protein is labeled with an arrowhead, and an unspecific band also present in untransfected cells is indicated by an asterisk. (B) HEK293T cells were transiently transfected with His6-FAT10, HA-Ub, HA-UbK48R, or HA-UbK48R/ΔGG. Prior to lysis, cells were incubated with 100 μM of the proteasome inhibitor LLnL for 6 h. After an immunoprecipitation (IP) with the anti-FAT10 antibody characterized in panel A, the precipitates were analyzed by Western blotting (WB) with either anti-His6 (left panel) or anti-HA (right panel) antibodies. (C) HEK293T cells were transiently transfected with constructs encoding FAT10-AV-GFP, ubiquitin-AV-GFP, SUMO-AV-GFP, UbK48R-AV-GFP, or GFP. Four hours before lysis, cells were treated with 50 μM of the proteasome inhibitor lactacystin (Lac) where indicated. Lysates were immunoprecipitated with anti-GFP antibody, and immunoprecipitates were analyzed by Western blotting with an antiubiquitin antibody. The arrowhead indicates the signal for Ub-AV-GFP and UbK48-AV-GFP, and polyubiquitin conjugates of the respective proteins are labeled poly-Ub-X-GFP.
FIG. 5.
Ubiquitylation of FAT10 is not necessary for FAT10 degradation. (A) FAT10, a lysineless FAT10 mutant (FAT10-K0), and NUB1L were transiently expressed as HA-tagged proteins in HEK293T cells and treated with 50 μM of lactacystin (Lac) during the labeling and the chase where indicated. The cells were labeled for 1 h with [35S]Cys-Met and chased for the indicated time periods prior to HA-specific immunoprecipitation, SDS/PAGE, and autoradiography. (B) HEK293T cells were transfected with HA-FAT10, HA-FAT10-K0, and HA-NUB1L as indicated on the top. After labeling, lysates were immunoprecipitated (IP) with anti-FAT10 or anti-HA antibodies as indicated and analyzed by SDS/PAGE and autoradiography. (C) HEK293T cells were transfected with HA-FAT10, HA-FAT10-K0, and HA-ubiquitin as indicated above. Four hours before lysis, cells were treated with 50 μM of lactacystin where indicated. As shown in the upper panel, lysates were immunoprecipitated with anti-FAT10 antibody, and immunoprecipitates were analyzed by Western blotting with anti-HA antibody. The lower panel shows an anti-HA Western blot of total lysates.
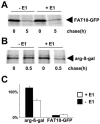
FIG. 6.
Degradation of FAT10 is ubiquitin independent. (A) The E1 thermosensitive cell line E36-ts20 (−E1) and E36-ts20 cells retransfected with a wild-type E1 (+E1) were transiently transfected with HA-tagged FAT10-AV-GFP. After inactivation of the thermolabile E1 by temperature shifting, the cells were labeled for 1 h with [35S]Cys-Met and chased for the indicated time periods prior to HA-specific immunoprecipitation, SDS/PAGE, and autoradiography. (B) Similar experiments were performed transfecting a short-lived HA-DHFR-ubiquitin-Arg-β-Galactosidase construct. The immunoprecipitation was performed with an anti-β-Gal antibody. (C) The bands were quantified on a radioimager and plotted as percent radioactivity based on values obtained directly after the pulse. The means of results from three independent experiments are shown, and error bars indicate the standard errors of the means.
Similar articles
- Degradation of FAT10 by the 26S proteasome is independent of ubiquitylation but relies on NUB1L.
Schmidtke G, Kalveram B, Groettrup M. Schmidtke G, et al. FEBS Lett. 2009 Feb 4;583(3):591-4. doi: 10.1016/j.febslet.2009.01.006. Epub 2009 Jan 21. FEBS Lett. 2009. PMID: 19166848 - FAT10ylation as a signal for proteasomal degradation.
Schmidtke G, Aichem A, Groettrup M. Schmidtke G, et al. Biochim Biophys Acta. 2014 Jan;1843(1):97-102. doi: 10.1016/j.bbamcr.2013.01.009. Epub 2013 Jan 18. Biochim Biophys Acta. 2014. PMID: 23333871 Review. - NEDD8 ultimate buster-1L interacts with the ubiquitin-like protein FAT10 and accelerates its degradation.
Hipp MS, Raasi S, Groettrup M, Schmidtke G. Hipp MS, et al. J Biol Chem. 2004 Apr 16;279(16):16503-10. doi: 10.1074/jbc.M310114200. Epub 2004 Feb 2. J Biol Chem. 2004. PMID: 14757770 - The ubiquitin-like modifier FAT10 interacts with HDAC6 and localizes to aggresomes under proteasome inhibition.
Kalveram B, Schmidtke G, Groettrup M. Kalveram B, et al. J Cell Sci. 2008 Dec 15;121(Pt 24):4079-88. doi: 10.1242/jcs.035006. Epub 2008 Nov 25. J Cell Sci. 2008. PMID: 19033385 - The ubiquitin-like modifier FAT10 in antigen processing and antimicrobial defense.
Basler M, Buerger S, Groettrup M. Basler M, et al. Mol Immunol. 2015 Dec;68(2 Pt A):129-32. doi: 10.1016/j.molimm.2015.04.012. Epub 2015 May 14. Mol Immunol. 2015. PMID: 25983082 Review.
Cited by
- Conjugation of the ubiquitin activating enzyme UBE1 with the ubiquitin-like modifier FAT10 targets it for proteasomal degradation.
Bialas J, Groettrup M, Aichem A. Bialas J, et al. PLoS One. 2015 Mar 13;10(3):e0120329. doi: 10.1371/journal.pone.0120329. eCollection 2015. PLoS One. 2015. PMID: 25768649 Free PMC article. - Infected erythrocytes and plasma proteomics reveal a specific protein signature of severe malaria.
Fraering J, Salnot V, Gautier EF, Ezinmegnon S, Argy N, Peoc'h K, Manceau H, Alao J, Guillonneau F, Migot-Nabias F, Bertin GI, Kamaliddin C; NeuroCM consortium. Fraering J, et al. EMBO Mol Med. 2024 Feb;16(2):319-333. doi: 10.1038/s44321-023-00010-0. Epub 2024 Jan 31. EMBO Mol Med. 2024. PMID: 38297098 Free PMC article. - ISG15 and immune diseases.
Jeon YJ, Yoo HM, Chung CH. Jeon YJ, et al. Biochim Biophys Acta. 2010 May;1802(5):485-96. doi: 10.1016/j.bbadis.2010.02.006. Epub 2010 Feb 12. Biochim Biophys Acta. 2010. PMID: 20153823 Free PMC article. Review. - FAT10 Induces cancer cell migration by stabilizing phosphorylated ABI3/NESH.
Um H, Jeong H, Lee B, Kim Y, Lee J, Roh JS, Lee SG, Park HR, Robinson WH, Sohn DH. Um H, et al. Anim Cells Syst (Seoul). 2023 Mar 11;27(1):53-60. doi: 10.1080/19768354.2023.2186486. eCollection 2023. Anim Cells Syst (Seoul). 2023. PMID: 36926204 Free PMC article. - The ubiquitin system: an essential component to unlocking the secrets of malaria parasite biology.
Hamilton MJ, Lee M, Le Roch KG. Hamilton MJ, et al. Mol Biosyst. 2014 Apr;10(4):715-23. doi: 10.1039/c3mb70506d. Epub 2014 Jan 30. Mol Biosyst. 2014. PMID: 24481176 Free PMC article. Review.
References
- Bates, E. F. M., O. Ravel, M. C. Dieu, S. Ho, C. Guret, J. M. Bridon, S. Ait-Yahia, F. Briere, C. Caux, J. Banchereau, and S. Lebecque. 1997. Identification and analysis of a novel member of the ubiquitin family expressed in dendritic cells and mature B cells. Eur. J. Immunol. 27:2471-2477. - PubMed
- Ciechanover, A., and R. Ben-Saadon. 2004. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14:103-106. - PubMed
- Elsasser, S., R. R. Gali, M. Schwickart, C. N. Larsen, D. S. Leggett, B. Muller, M. T. Feng, F. Tubing, G. A. G. Dittmar, and D. Finley. 2002. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4:725-730. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases