Identification in vivo of different rate-limiting steps associated with transcriptional activators in the presence and absence of a GAGA element - PubMed (original) (raw)
Identification in vivo of different rate-limiting steps associated with transcriptional activators in the presence and absence of a GAGA element
Yunyuan Vivian Wang et al. Mol Cell Biol. 2005 May.
Abstract
We analyzed the impact of a GAGA element on a transgenic promoter in Drosophila melanogaster that was activated by proteins composed of the Tet(on) DNA binding domain and either the heat shock factor (HSF) activation domain or a potent subdomain of VP16. Permanganate footprinting was used to monitor polymerase II (Pol II) on the transgenic promoters in vivo. Activation by Tet(on)-HSF but not by Tet(on)-VP16(A2) required the GAGA element; this correlated with the ability of the GAGA element to establish a paused Pol II. Although the GAGA element was not required for activation by Tet(on)-VP16(A2), the GAGA element greatly accelerated the rate of activation. The permanganate data also provided evidence that Pol II encountered different rate-limiting steps, following initiation in the presence of Tet(on)-HSF and Tet(on)-VP16(A2). The rate-limiting step in the presence of Tet(on)-HSF was release of Pol II paused about 20 to 40 nucleotides downstream from the start site. The rate-limiting step in the presence of Tet(on)-VP16(A2) occurred much closer to the transcription start site. Several biochemical studies have provided evidence for a structural transition shortly after Pol II initiates transcription. The behavior of Pol II in the presence of Tet(on)-VP16(A2) provides the first evidence that this transition occurs in vivo.
Figures
FIG. 1.
The HSF activation domain requires a GAGA element in the target promoter to achieve activation. Transgenic fly lines encoding Teton-HSF (left, A to D), Teton-VP16A2 (E and F), or Teton-DBD (G and H) were crossed with transgenic lines containing TRE5 (A, C, E, and G) or TRE5-GA (B, D, F, and H) promoters. The offspring were raised in the presence or absence of doxycycline (+ Dox, −Dox), heat shocked for 30 min to induce synthesis of Teton proteins, and allowed to recover overnight at 22°C. Salivary glands (yellow arrowheads) and fat body (red arrowheads) were isolated and incubated in X-Gal solution. Tissues expressing Teton-HSF or Teton-DBD were incubated with X-Gal for 5 h, and tissues expressing Teton-VP16A2 were incubated for 30 min. (Right) Schematics of the transgenic target genes and transgenic activators used in this study. The wild type (WT) is a transgene that has the hsp70 promoter from −194 to +84 positioned upstream from sequences encoding β-galactosidase. GAGA elements are green, heat-shock elements (HSE) are pink, the hsp70 core promoter region spanning −44 to +84 is light blue, and the β-galactosidase sequence is dark blue. TRE5 has five binding sites for Teton (grey boxes) located upstream from the hsp70 core promoter. TRE5-GA has a GAGA element inserted between the core promoter and the Tet upstream activation sequence (UAS). All three of the transgenic activators share the Teton DBD. Teton-VP16A2 has an activation domain consisting of three tandem copies of a 13-amino-acid peptide derived from VP16 (2). Teton-HSF has an activation domain consisting of the last 81 amino acids of Drosophila HSF. Teton-DBD lacks an activation domain.
FIG. 2.
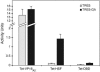
Measurement of β-galactosidase activity by various Teton-fusion proteins. Transgenic fly lines encoding Teton-VP16A2, Teton-HSF, and Teton-DBD were crossed with transgenic lines containing TRE5 or TRE5-GA promoters. The offspring were raised in the presence of doxycycline, heat shocked for 30 min to induce synthesis of Teton proteins, and allowed to recover for 3 h. Fat bodies from five larvae were collected for the chlorophenol red-beta-
d
-galactopyranoside assay. β-Galactosidase activity was defined as absorbance units per hour per milligram of protein.
FIG. 3.
Permanganate footprinting analysis of TRE5 and TRE5-GA target genes subjected to the action of various Teton fusion proteins. (A) Fly lines containing transgenes for Teton-VP16A2 (lanes 1 through 4), Teton-HSF (lanes 5 through 8), or Teton-DBD (lanes 9 through 12) were crossed with fly lines containing the TRE5 (lanes 1, 2, 5, 6, 9, and 10) or TRE5-GA (lanes 3, 4, 7, 8, 11, and 12) targets. Offspring were raised either in the absence (odd-numbered lanes) or presence (even-numbered lanes) of doxycycline. Larvae were heat shocked for 30 min, followed by a 1-h recovery to induce synthesis of the Teton proteins. Fat bodies were then subjected to permanganate genomic footprinting. (B) Permanganate footprinting of transgenic targets in fat bodies. See the legend to Fig. 1 for a description of the target genes WT, TRE5, and TRE5-GA. No Teton derivatives were present in these tissues. (C) Western blot analysis of various Teton-fusion proteins produced in fat bodies. Fat bodies from five larvae (lanes 1, 3, 5, and 7) or one-third of this amount (lanes 2, 4, 6, and 8) were run on the gel. Teton fusion proteins (open arrowheads) were detected with antibody raised against the Teton-DBD. The two bands marked by asterisks represent background proteins detected with this antibody, and they serve as loading controls.
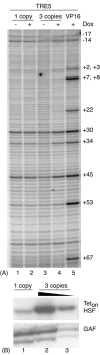
FIG. 4.
Activation of TRE5 cannot be achieved by increasing the amount of Teton-HSF in fat bodies. (A) Fat bodies were isolated from larvae containing one copy of the TRE5 transgene and either one or three copies of the transgene for Teton-HSF or one copy of the transgene for Teton-VP16A2. Larvae were raised either in the absence (lanes 1 and 3) or presence (lanes 2, 4, and 5) of doxycycline. Fat bodies were subjected to permanganate footprinting. (B) Western blot analysis of Teton-HSF proteins produced in fat bodies. Teton-HSF was detected in extracts from the transgenic flies that contain one copy (lane 1) or three copies (lane 2, one-third dilution in lane 3) of Teton-HSF transgene (with antibody against Teton-DBD). Samples were also analyzed for the presence of GAGA factor (GAF) as a control for sample loading.
FIG. 5.
Kinetic analysis of activation in fat bodies. Fly lines containing transgenes for Teton-HSF or Teton-VP16A2 were crossed with fly lines containing the TRE5 or the TRE5-GA transgenes. Larvae were raised in the absence of doxycycline. Fat bodies were isolated from larvae, following production of the Teton proteins in larvae by heat shock and recovery. The fat bodies were incubated in medium containing 5 μg of doxycycline/ml for 0, 5, 10, 30, or 60 min, followed by permanganate treatment. Lanes 1 to 5, permanganate reactivity of TRE5-GA at various times following the induction of binding of Teton-HSF; lanes 7 to 11, permanganate reactivity of TRE5 at various times following the induction of binding of Teton-VP16A2; lanes 13 to 17, permanganate reactivity of TRE5-GA at various times following the induction of binding of Teton-VP16A2. Lanes 6, 12, and 18 show permanganate sensitivity of target genes in fat bodies when larvae were fed doxycycline.
FIG. 6.
Kinetic analysis of TRE5-GA activation by Teton-VP16A2 in salivary glands. (A) Permanganate footprinting of transgenic targets in salivary glands. See the legend to Fig. 1 for a description of the target genes WT, TRE5, and TRE5-GA. (B) Fly lines expressing Teton-VP16A2 were crossed with fly lines containing TRE5-GA. Larvae were raised in the absence of doxycycline. Production of Teton-VP16A2 was induced by 30-min heat shock, followed by 1-h recovery at room temperature. Salivary glands were isolated from larvae and subjected to doxycycline induction and permanganate footprinting as described for fat bodies.
Similar articles
- HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites.
Shopland LS, Hirayoshi K, Fernandes M, Lis JT. Shopland LS, et al. Genes Dev. 1995 Nov 15;9(22):2756-69. doi: 10.1101/gad.9.22.2756. Genes Dev. 1995. PMID: 7590251 - Cooperative and competitive protein interactions at the hsp70 promoter.
Mason PB Jr, Lis JT. Mason PB Jr, et al. J Biol Chem. 1997 Dec 26;272(52):33227-33. doi: 10.1074/jbc.272.52.33227. J Biol Chem. 1997. PMID: 9407112 - NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila.
Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. Lee C, et al. Mol Cell Biol. 2008 May;28(10):3290-300. doi: 10.1128/MCB.02224-07. Epub 2008 Mar 10. Mol Cell Biol. 2008. PMID: 18332113 Free PMC article. - The heat shock response: A case study of chromatin dynamics in gene regulation.
Teves SS, Henikoff S. Teves SS, et al. Biochem Cell Biol. 2013 Feb;91(1):42-8. doi: 10.1139/bcb-2012-0075. Epub 2013 Feb 13. Biochem Cell Biol. 2013. PMID: 23442140 Review. - Transcriptional activation: is it rocket science?
Pollock R, Gilman M. Pollock R, et al. Proc Natl Acad Sci U S A. 1997 Dec 9;94(25):13388-9. doi: 10.1073/pnas.94.25.13388. Proc Natl Acad Sci U S A. 1997. PMID: 9391033 Free PMC article. Review. No abstract available.
Cited by
- Promoter elements associated with RNA Pol II stalling in the Drosophila embryo.
Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. Hendrix DA, et al. Proc Natl Acad Sci U S A. 2008 Jun 3;105(22):7762-7. doi: 10.1073/pnas.0802406105. Epub 2008 May 27. Proc Natl Acad Sci U S A. 2008. PMID: 18505835 Free PMC article. - Transcription regulation through promoter-proximal pausing of RNA polymerase II.
Core LJ, Lis JT. Core LJ, et al. Science. 2008 Mar 28;319(5871):1791-2. doi: 10.1126/science.1150843. Science. 2008. PMID: 18369138 Free PMC article. - GAGA factor: a multifunctional pioneering chromatin protein.
Chetverina D, Erokhin M, Schedl P. Chetverina D, et al. Cell Mol Life Sci. 2021 May;78(9):4125-4141. doi: 10.1007/s00018-021-03776-z. Epub 2021 Feb 2. Cell Mol Life Sci. 2021. PMID: 33528710 Free PMC article. Review. - Activity of heat shock genes' promoters in thermally contrasting animal species.
Astakhova LN, Zatsepina OG, Funikov SY, Zelentsova ES, Schostak NG, Orishchenko KE, Evgen'ev MB, Garbuz DG. Astakhova LN, et al. PLoS One. 2015 Feb 20;10(2):e0115536. doi: 10.1371/journal.pone.0115536. eCollection 2015. PLoS One. 2015. PMID: 25700087 Free PMC article. - Promoter proximal pausing on genes in metazoans.
Gilmour DS. Gilmour DS. Chromosoma. 2009 Feb;118(1):1-10. doi: 10.1007/s00412-008-0182-4. Epub 2008 Oct 2. Chromosoma. 2009. PMID: 18830703 Review.
References
- Chen, H. T., and S. Hahn. 2004. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell 119:169-180. - PubMed
- Clos, J., J. T. Westwood, P. B. Becker, S. Wilson, K. Lambert, and C. Wu. 1990. Molecular cloning and expression of a hexameric Drosophila heat shock factor subject to negative regulation. Cell 63:1085-1097. - PubMed
- Dellino, G. I., Y. B. Schwartz, G. Farkas, D. McCabe, S. C. Elgin, and V. Pirrotta. 2004. Polycomb silencing blocks transcription initiation. Mol. Cell 13:887-893. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases