The genome of S-PM2, a "photosynthetic" T4-type bacteriophage that infects marine Synechococcus strains - PubMed (original) (raw)
The genome of S-PM2, a "photosynthetic" T4-type bacteriophage that infects marine Synechococcus strains
Nicholas H Mann et al. J Bacteriol. 2005 May.
Abstract
Bacteriophage S-PM2 infects several strains of the abundant and ecologically important marine cyanobacterium Synechococcus. A large lytic phage with an isometric icosahedral head, S-PM2 has a contractile tail and by this criterion is classified as a myovirus (1). The linear, circularly permuted, 196,280-bp double-stranded DNA genome of S-PM2 contains 37.8% G+C residues. It encodes 239 open reading frames (ORFs) and 25 tRNAs. Of these ORFs, 19 appear to encode proteins associated with the cell envelope, including a putative S-layer-associated protein. Twenty additional S-PM2 ORFs have homologues in the genomes of their cyanobacterial hosts. There is a group I self-splicing intron within the gene encoding the D1 protein. A total of 40 ORFs, organized into discrete clusters, encode homologues of T4 proteins involved in virion morphogenesis, nucleotide metabolism, gene regulation, and DNA replication and repair. The S-PM2 genome encodes a few surprisingly large (e.g., 3,779 amino acids) ORFs of unknown function. Our analysis of the S-PM2 genome suggests that many of the unknown S-PM2 functions may be involved in the adaptation of the metabolism of the host cell to the requirements of phage infection. This hypothesis originates from the identification of multiple phage-mediated modifications of the host's photosynthetic apparatus that appear to be essential for maintaining energy production during the lytic cycle.
Figures
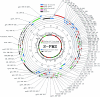
FIG. 1.
Organization of the genome of phage S-PM2. The circles from outside to inside indicate the following: 1 to 6 represent the six reading frames, 7 is the scale bar (in kilobases), 8 is G+C content (smoothed with a sigma = 200 bp; Gaussian; range is 28.2 to 60.4%, with a mean of 37.8%), and 9 shows homology with other organisms. Labels show ORF numbers and gene designations where a putative homologue in T4 has been identified. MS indicates that presence of protein in the virion has been established by mass spectrometry and is shown in parentheses if the ORF has already been shown to encode a virion structural protein on the basis of proposed homology. The following color scheme has been used: green, ORFs encoding proteins exhibiting similarity to cyanobacterial proteins; blue, phage structural proteins; red, other phage proteins; purple, tRNA genes; and black, unidentified ORFs. The four clusters of T4-like genes are shaded in gray.
FIG. 2.
Consensus features of S-PM2 putative early (A) and late (B) promoters. The features were calculated by using WebLogo (12). The height of each stack indicates the sequence conservation at that position (measured in bits), whereas the height of each symbol within the stack reflects the relative frequency of the corresponding base at that position. The consensus promoter features of phages T4 and RB49 (15) are shown for comparison.
Similar articles
- Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus.
Weigele PR, Pope WH, Pedulla ML, Houtz JM, Smith AL, Conway JF, King J, Hatfull GF, Lawrence JG, Hendrix RW. Weigele PR, et al. Environ Microbiol. 2007 Jul;9(7):1675-95. doi: 10.1111/j.1462-2920.2007.01285.x. Environ Microbiol. 2007. PMID: 17564603 - Molecular Analysis of Arthrobacter Myovirus vB_ArtM-ArV1: We Blame It on the Tail.
Kaliniene L, Šimoliūnas E, Truncaitė L, Zajančkauskaitė A, Nainys J, Kaupinis A, Valius M, Meškys R. Kaliniene L, et al. J Virol. 2017 Mar 29;91(8):e00023-17. doi: 10.1128/JVI.00023-17. Print 2017 Apr 15. J Virol. 2017. PMID: 28122988 Free PMC article. - A proteomic approach to the identification of the major virion structural proteins of the marine cyanomyovirus S-PM2.
Clokie MRJ, Thalassinos K, Boulanger P, Slade SE, Stoilova-McPhie S, Cane M, Scrivens JH, Mann NH. Clokie MRJ, et al. Microbiology (Reading). 2008 Jun;154(Pt 6):1775-1782. doi: 10.1099/mic.0.2007/016261-0. Microbiology (Reading). 2008. PMID: 18524932 - Sequence analysis of Leuconostoc mesenteroides bacteriophage Phi1-A4 isolated from an industrial vegetable fermentation.
Lu Z, Altermann E, Breidt F, Kozyavkin S. Lu Z, et al. Appl Environ Microbiol. 2010 Mar;76(6):1955-66. doi: 10.1128/AEM.02126-09. Epub 2010 Jan 29. Appl Environ Microbiol. 2010. PMID: 20118355 Free PMC article. - Cyanophage infection and photoinhibition in marine cyanobacteria.
Bailey S, Clokie MR, Millard A, Mann NH. Bailey S, et al. Res Microbiol. 2004 Nov;155(9):720-5. doi: 10.1016/j.resmic.2004.06.002. Res Microbiol. 2004. PMID: 15501648 Review.
Cited by
- Genomic characteristics and environmental distributions of the uncultivated Far-T4 phages.
Roux S, Enault F, Ravet V, Pereira O, Sullivan MB. Roux S, et al. Front Microbiol. 2015 Mar 16;6:199. doi: 10.3389/fmicb.2015.00199. eCollection 2015. Front Microbiol. 2015. PMID: 25852662 Free PMC article. - Virus-host protein-protein interactions of mycobacteriophage Giles.
Mehla J, Dedrick RM, Caufield JH, Wagemans J, Sakhawalkar N, Johnson A, Hatfull GF, Uetz P. Mehla J, et al. Sci Rep. 2017 Nov 28;7(1):16514. doi: 10.1038/s41598-017-16303-7. Sci Rep. 2017. PMID: 29184079 Free PMC article. - Transcriptome dynamics of a broad host-range cyanophage and its hosts.
Doron S, Fedida A, Hernández-Prieto MA, Sabehi G, Karunker I, Stazic D, Feingersch R, Steglich C, Futschik M, Lindell D, Sorek R. Doron S, et al. ISME J. 2016 Jun;10(6):1437-55. doi: 10.1038/ismej.2015.210. Epub 2015 Dec 1. ISME J. 2016. PMID: 26623542 Free PMC article. - Marine phage genomics: the tip of the iceberg.
Perez Sepulveda B, Redgwell T, Rihtman B, Pitt F, Scanlan DJ, Millard A. Perez Sepulveda B, et al. FEMS Microbiol Lett. 2016 Aug;363(15):fnw158. doi: 10.1093/femsle/fnw158. Epub 2016 Jun 22. FEMS Microbiol Lett. 2016. PMID: 27338950 Free PMC article. Review. - Regulation of infection efficiency in a globally abundant marine Bacteriodetes virus.
Howard-Varona C, Roux S, Dore H, Solonenko NE, Holmfeldt K, Markillie LM, Orr G, Sullivan MB. Howard-Varona C, et al. ISME J. 2017 Jan;11(1):284-295. doi: 10.1038/ismej.2016.81. Epub 2016 May 17. ISME J. 2017. PMID: 27187794 Free PMC article.
References
- Ackermann, H. W., and M. S. DuBow. 1987. Viruses of prokaryotes. Bacteriophage taxonomy, p. 13-44. CRC Press, Boca Raton, Fla.
- Bhaya, D., A. Dufresne, D. Vaulot, and A. Grossman. 2002. Analysis of the hli gene family in marine and freshwater cyanobacteria. FEMS Microbiol. Lett. 215:209-219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources