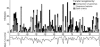Examining bacterial species under the specter of gene transfer and exchange - PubMed (original) (raw)
Review
. 2005 May 3;102 Suppl 1(Suppl 1):6595-9.
doi: 10.1073/pnas.0502035102. Epub 2005 Apr 25.
Affiliations
- PMID: 15851673
- PMCID: PMC1131874
- DOI: 10.1073/pnas.0502035102
Review
Examining bacterial species under the specter of gene transfer and exchange
Howard Ochman et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Even in lieu of a dependable species concept for asexual organisms, the classification of bacteria into discrete taxonomic units is considered to be obstructed by the potential for lateral gene transfer (LGT) among lineages at virtually all phylogenetic levels. In most bacterial genomes, large proportions of genes are introduced by LGT, as indicated by their compositional features and/or phylogenetic distributions, and there is also clear evidence of LGT between very distantly related organisms. By adopting a whole-genome approach, which examined the history of every gene in numerous bacterial genomes, we show that LGT does not hamper phylogenetic reconstruction at many of the shallower taxonomic levels. Despite the high levels of gene acquisition, the only taxonomic group for which appreciable amounts of homologous recombination were detected was within bacterial species. Taken as a whole, the results derived from the analysis of complete gene inventories support several of the current means to recognize and define bacterial species.
Figures
Fig. 1.
Linear representation of the E. coli MG1655 chromosome showing the distribution of horizontally acquired DNA. At each minute (1-100), vertical bars depict the amount of horizontally acquired, protein-coding DNA, as inferred by two methods: (i) atypical sequence features (i.e., base composition) (white) and (ii) the unique occurrence of a gene in E. coli after aligning and comparing the genome sequences of E. coli, Salmonella enterica, and Klebsiella pneumonia (black). Gray portions of vertical bars denote the overlap between the methods and the amount of protein-coding DNA in E. coli inferred to be horizontally acquired based both on its sequence features and on its phylogenetic distribution. Along the bottom of the figure is shown the base composition (% G+C) computed in discrete windows for each minute of the chromosome. The dashed horizontal line shows the overall average base composition for all protein-coding genes in this genome (51.0% G+C). Figure modified from Lawrence and Ochman (18).
Fig. 2.
Phylogenetic inference of cohesion within bacterial genomes. (A) Relationship between gene acquisition and loss and the amount of phylogenetic incongruence observed in fully sequenced bacterial genomes. For each quartet of genomes, we inferred the number of recently acquired and lost genes (shown at arrows on the corresponding branches). In addition, for each quartet of genomes, orthologous genes were inferred, aligned, and evaluated at the nucleic sequence level based on the Shimodaira-Hasegawa test implemented in Puzzle 5.1 (27, 28). Shown are the numbers of orthologs supporting each category of alignment: (i) those supporting the reference phylogeny (rRNA topology), (ii) those supporting either alternate phylogeny (LGT topology), and (iii) those with no statistical support for any phylogeny (nonresolving). (B) Relative frequencies of the three categories of alignments in diverse bacterial groups at several taxonomic levels. The shaded zone of this plot represents the area where LGT predominates. Only for intraspecies comparisons within E. coli and C. pneumoniae are the frequencies of LGT >5%. Data and figure from Daubin et al. (26).
Fig. 3.
Example of a lateral gene transfer detected by phylogenetic discordance. (A) Neighbor-joining tree based on the concatenation of 205 single-copy genes common to all 13 Gammaproteobacterial species. Note that 203 of the 205 genes individually supported the same topology. Figure adapted from Lerat et al. (38). (B) Example of a tree that conflicts with the reference topology. Among the small proportion of proteins showing statistical support for an alternate topology was tellurium resistance protein. Homologs of the gene encoding this protein have been detected in only 9 of the 13 species. Note that the topology of this tree departs from that of the reference tree (depicted in A) because of a single LGT event that occurred in the ancestor to E. coli and Salmonella enterica. Data in B are from Lerat et al. (37).
Similar articles
- Phylogenomic networks.
Dagan T. Dagan T. Trends Microbiol. 2011 Oct;19(10):483-91. doi: 10.1016/j.tim.2011.07.001. Epub 2011 Aug 3. Trends Microbiol. 2011. PMID: 21820313 Review. - Lateral Gene Transfer Dynamics in the Ancient Bacterial Genus Streptomyces.
McDonald BR, Currie CR. McDonald BR, et al. mBio. 2017 Jun 6;8(3):e00644-17. doi: 10.1128/mBio.00644-17. mBio. 2017. PMID: 28588130 Free PMC article. - Lateral gene transfer as a support for the tree of life.
Abby SS, Tannier E, Gouy M, Daubin V. Abby SS, et al. Proc Natl Acad Sci U S A. 2012 Mar 27;109(13):4962-7. doi: 10.1073/pnas.1116871109. Epub 2012 Mar 13. Proc Natl Acad Sci U S A. 2012. PMID: 22416123 Free PMC article. - Recovering the treelike trend of evolution despite extensive lateral genetic transfer: a probabilistic analysis.
Roch S, Snir S. Roch S, et al. J Comput Biol. 2013 Feb;20(2):93-112. doi: 10.1089/cmb.2012.0234. J Comput Biol. 2013. PMID: 23383996 Free PMC article. - [Homologous recombination among bacterial genomes: the measurement and identification].
Yang XW, Yang RF, Cui YJ. Yang XW, et al. Yi Chuan. 2016 Feb;38(2):137-43. doi: 10.16288/j.yczz.15-382. Yi Chuan. 2016. PMID: 26907777 Review. Chinese.
Cited by
- Cassette recruitment in the chromosomal Integron of Vibrio cholerae.
Vit C, Richard E, Fournes F, Whiteway C, Eyer X, Lapaillerie D, Parissi V, Mazel D, Loot C. Vit C, et al. Nucleic Acids Res. 2021 Jun 4;49(10):5654-5670. doi: 10.1093/nar/gkab412. Nucleic Acids Res. 2021. PMID: 34048565 Free PMC article. - Genome-wide detection and analysis of homologous recombination among sequenced strains of Escherichia coli.
Mau B, Glasner JD, Darling AE, Perna NT. Mau B, et al. Genome Biol. 2006;7(5):R44. doi: 10.1186/gb-2006-7-5-r44. Epub 2006 May 31. Genome Biol. 2006. PMID: 16737554 Free PMC article. - Gene Transmission in the One Health Microbiosphere and the Channels of Antimicrobial Resistance.
Baquero F, Coque TM, Martínez JL, Aracil-Gisbert S, Lanza VF. Baquero F, et al. Front Microbiol. 2019 Dec 17;10:2892. doi: 10.3389/fmicb.2019.02892. eCollection 2019. Front Microbiol. 2019. PMID: 31921068 Free PMC article. Review. - Gene and genome trees conflict at many levels.
Haggerty LS, Martin FJ, Fitzpatrick DA, McInerney JO. Haggerty LS, et al. Philos Trans R Soc Lond B Biol Sci. 2009 Aug 12;364(1527):2209-19. doi: 10.1098/rstb.2009.0042. Philos Trans R Soc Lond B Biol Sci. 2009. PMID: 19571241 Free PMC article. - The extent of genome flux and its role in the differentiation of bacterial lineages.
Nowell RW, Green S, Laue BE, Sharp PM. Nowell RW, et al. Genome Biol Evol. 2014 Jun 12;6(6):1514-29. doi: 10.1093/gbe/evu123. Genome Biol Evol. 2014. PMID: 24923323 Free PMC article.
References
- Rossello-Mora, R. & Amann, R. (2001) FEMS Microbiol. Rev. 25, 39-67. - PubMed
- Lan, R. & Reeves, P. R. (2001) Trends Microbiol. 9, 419-424. - PubMed
- Young, J. M. (2001) Int. J. Syst. Evol. Microbiol. 51, 945-953. - PubMed
- Cohan, F. M. (2002) Annu. Rev. Microbiol. 56, 457-487. - PubMed
- Ochman, H., Lawrence, J. G. & Groisman, E. A. (2000) Nature 405, 299-304. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources