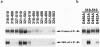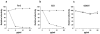Evaluation of human monoclonal antibody 80R for immunoprophylaxis of severe acute respiratory syndrome by an animal study, epitope mapping, and analysis of spike variants - PubMed (original) (raw)
Evaluation of human monoclonal antibody 80R for immunoprophylaxis of severe acute respiratory syndrome by an animal study, epitope mapping, and analysis of spike variants
Jianhua Sui et al. J Virol. 2005 May.
Abstract
In this report, the antiviral activity of 80R immunoglobulin G1 (IgG1), a human monoclonal antibody against severe acute respiratory syndrome coronavirus (SARS-CoV) spike (S) protein that acts as a viral entry inhibitor in vitro, was investigated in vivo in a mouse model. When 80R IgG1 was given prophylactically to mice at doses therapeutically achievable in humans, viral replication was reduced by more than 4 orders of magnitude to below assay limits. The essential core region of S protein required for 80R binding was identified as a conformationally sensitive fragment (residues 324 to 503) that overlaps the receptor ACE2-binding domain. Amino acids critical for 80R binding were identified. In addition, the effects of various 80R-binding domain amino acid substitutions which occur in SARS-like-CoV from civet cats, and which evolved during the 2002/2003 outbreak and in a 2003/2004 Guangdong index patient, were analyzed. The results demonstrated that the vast majority of SARS-CoVs are sensitive to 80R. We propose that by establishing the susceptibility and resistance profiles of newly emerging SARS-CoVs through early S1 genotyping of the core 180-amino-acid neutralizing epitope of 80R, an effective immunoprophylaxis strategy with 80R should be possible in an outbreak setting. Our study also cautions that for any prophylaxis strategy based on neutralizing antibody responses, whether by passive or active immunization, a genotyping monitor will be necessary for effective use.
Figures
FIG. 1.
Truncations and point mutations of S1(318-510) were analyzed to define the 80R antibody epitope. S1 residues 318 to 510 fused to the Fc region of human IgG1 and truncation or mutation variants of S1(318-510) containing the indicated residues were metabolically labeled and precipitated by protein A or 80R scFv. (a) S1(324-503) was the smallest fragment bound to 80R. Either N-terminal or C-terminal truncation variants slightly smaller than that either had decreased expression or lost binding activity to 80R. (b) Critical residues for the 80R epitope were observed. Individual alanine substitution of glutamic acid 452 or aspartic acids 454 and 480 in the S1(318-510) fragment impaired or abolished binding to 80R. IP, immunoprecipitation.
FIG. 2.
Effects on 80R binding of variant amino acid substitutions of S protein that occur in animal SARS-like-CoVs and human SARS-CoVs. (a) Indicated amino acid residues in S1(318-510) of Tor2 were individually replaced with corresponding variant amino acids found in SARS-like-CoVs or other human SARS-CoVs. These residues also were replaced with alanine. Alterations of K344A, N479A, and T487A affected the binding to 80R to some degree. N479K substitution resulted in an ∼50% decrease in 80R binding, and D480G substitution totally abolished binding to 80R. (b) Multiple substitutions with the amino acids of civet SZ3 virus (344R/360S/479K/487S) in the S1(318-510)-Ig construct of Tor2 had no effect on 80R binding, as well as full-length S1(12-672) of SZ3, which was de novo synthesized. Multiple substitutions with the amino acids of human GD03T virus (344R/360S/472P/480G/487S) in the S1(318-510)-Ig construct of Tor2 and full-length S1(12-672) of GD03T completely lost binding to 80R scFv. (c) Full-length S protein of Tor2 and variants containing amino acid substitutions of isolates SZ3 or GD03T were precipitated by 1D4, which recognizes a C9 tag present at the carboxyl terminus of each S protein, or by 80R IgG1 and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The binding activities of these full-length S proteins to 80R IgG1 were consistent with those of their receptor-binding domains (318 to 510) or S1 domains (12 to 672) to 80R scFv. WT, wild type; IP, immunoprecipitation.
FIG. 3.
80R IgG1 neutralization of pseudoviral infection mediated by full-length SARS-CoV spike variants. HIVs pseudotyped with the S protein from Tor2, SZ3, or GD03T isolates were incubated with the indicated concentration of 80R IgG1 (solid line, diamonds) or nonrelevant human IgG1 (solid line, squares) for 1 h prior to infection. At 48 h after infection, luciferase activities in target cells were measured and relative viral inhibition was calculated as the ratio of luciferase activity in the presence to absence of 80R IgG1 or nonrelevant human IgG1. (a) 80R IgG1 efficiently blocked Tor2 S protein-pseudotyped HIV infection, with a 90% inhibitory concentration around 2 μg/ml. (b) 80R IgG1 also efficiently neutralized SZ3 S protein pseudoviral infection. (c) In contrast to Tor2 and SZ3, GD03T S protein-pseudotyped virus was essentially resistant to the neutralization of 80R IgG1 with a concentration up to 50 μg/ml. These results are representative of two experiments with similar results.
Similar articles
- An exposed domain in the severe acute respiratory syndrome coronavirus spike protein induces neutralizing antibodies.
Zhou T, Wang H, Luo D, Rowe T, Wang Z, Hogan RJ, Qiu S, Bunzel RJ, Huang G, Mishra V, Voss TG, Kimberly R, Luo M. Zhou T, et al. J Virol. 2004 Jul;78(13):7217-26. doi: 10.1128/JVI.78.13.7217-7226.2004. J Virol. 2004. PMID: 15194798 Free PMC article. - Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants.
ter Meulen J, van den Brink EN, Poon LL, Marissen WE, Leung CS, Cox F, Cheung CY, Bakker AQ, Bogaards JA, van Deventer E, Preiser W, Doerr HW, Chow VT, de Kruif J, Peiris JS, Goudsmit J. ter Meulen J, et al. PLoS Med. 2006 Jul;3(7):e237. doi: 10.1371/journal.pmed.0030237. PLoS Med. 2006. PMID: 16796401 Free PMC article. - Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region.
Chen Z, Zhang L, Qin C, Ba L, Yi CE, Zhang F, Wei Q, He T, Yu W, Yu J, Gao H, Tu X, Gettie A, Farzan M, Yuen KY, Ho DD. Chen Z, et al. J Virol. 2005 Mar;79(5):2678-88. doi: 10.1128/JVI.79.5.2678-2688.2005. J Virol. 2005. PMID: 15708987 Free PMC article. - Vaccine design for severe acute respiratory syndrome coronavirus.
He Y, Jiang S. He Y, et al. Viral Immunol. 2005;18(2):327-32. doi: 10.1089/vim.2005.18.327. Viral Immunol. 2005. PMID: 16035944 Review. - The spike protein of SARS-CoV--a target for vaccine and therapeutic development.
Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. Du L, et al. Nat Rev Microbiol. 2009 Mar;7(3):226-36. doi: 10.1038/nrmicro2090. Epub 2009 Feb 9. Nat Rev Microbiol. 2009. PMID: 19198616 Free PMC article. Review.
Cited by
- Broad-Spectrum Anti-coronavirus Vaccines and Therapeutics to Combat the Current COVID-19 Pandemic and Future Coronavirus Disease Outbreaks.
Cao M, Su X, Jiang S. Cao M, et al. Stem Cell Reports. 2021 Mar 9;16(3):398-411. doi: 10.1016/j.stemcr.2020.12.010. Stem Cell Reports. 2021. PMID: 33691145 Free PMC article. Review. - Computational characterization and design of SARS coronavirus receptor recognition and antibody neutralization.
Zhang Y, Zheng N, Zhong Y. Zhang Y, et al. Comput Biol Chem. 2007 Apr;31(2):129-33. doi: 10.1016/j.compbiolchem.2007.02.005. Epub 2007 Feb 17. Comput Biol Chem. 2007. PMID: 17374510 Free PMC article. - Molecular pathogenesis of severe acute respiratory syndrome.
Wang H, Rao S, Jiang C. Wang H, et al. Microbes Infect. 2007 Jan;9(1):119-26. doi: 10.1016/j.micinf.2006.06.012. Epub 2006 Sep 28. Microbes Infect. 2007. PMID: 17142081 Free PMC article. Review. - Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody.
Prabakaran P, Gan J, Feng Y, Zhu Z, Choudhry V, Xiao X, Ji X, Dimitrov DS. Prabakaran P, et al. J Biol Chem. 2006 Jun 9;281(23):15829-36. doi: 10.1074/jbc.M600697200. Epub 2006 Apr 5. J Biol Chem. 2006. PMID: 16597622 Free PMC article. - Chimeric coronavirus-like particles carrying severe acute respiratory syndrome coronavirus (SCoV) S protein protect mice against challenge with SCoV.
Lokugamage KG, Yoshikawa-Iwata N, Ito N, Watts DM, Wyde PR, Wang N, Newman P, Kent Tseng CT, Peters CJ, Makino S. Lokugamage KG, et al. Vaccine. 2008 Feb 6;26(6):797-808. doi: 10.1016/j.vaccine.2007.11.092. Epub 2007 Dec 26. Vaccine. 2008. PMID: 18191004 Free PMC article.
References
- Burnouf, T., and M. Radosevich. 2003. Treatment of severe acute respiratory syndrome with convalescent plasma. Hong Kong Med. J. 9:309-310. - PubMed
- Chinese SARS Molecular Epidemiology Consortium. 2004. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303:1666-1669. - PubMed
- Guan, Y. 2004. An averted SARS outbreak. Hong Kong SARS Forum and Hospital Authority Convention, Hong Kong Hospital Authority, Hong Kong.
- Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI061318/AI/NIAID NIH HHS/United States
- P30 AI028691/AI/NIAID NIH HHS/United States
- R01 AI048436/AI/NIAID NIH HHS/United States
- U01 AI061318/AI/NIAID NIH HHS/United States
- AI28785/AI/NIAID NIH HHS/United States
- AI053822/AI/NIAID NIH HHS/United States
- AI48436/AI/NIAID NIH HHS/United States
- AI28691/AI/NIAID NIH HHS/United States
- R21 AI053822/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous