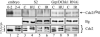Grp/DChk1 is required for G2-M checkpoint activation in Drosophila S2 cells, whereas Dmnk/DChk2 is dispensable - PubMed (original) (raw)
Grp/DChk1 is required for G2-M checkpoint activation in Drosophila S2 cells, whereas Dmnk/DChk2 is dispensable
Hilda I de Vries et al. J Cell Sci. 2005.
Abstract
Cell-cycle checkpoints are signal-transduction pathways required to maintain genomic stability in dividing cells. Previously, it was reported that two kinases essential for checkpoint signalling, Chk1 and Chk2 are structurally conserved. In contrast to yeast, Xenopus and mammals, the Chk1- and Chk2-dependent pathways in Drosophila are not understood in detail. Here, we report the function of these checkpoint kinases, referred to as Grp/DChk1 and Dmnk/DChk2 in Drosophila Schneider's cells, and identify an upstream regulator as well as downstream targets of Grp/DChk1. First, we demonstrate that S2 cells are a suitable model for G(2)/M checkpoint studies. S2 cells display Grp/DChk1-dependent and Dmnk/DChk2-independent cell-cycle-checkpoint activation in response to hydroxyurea and ionizing radiation. S2 cells depleted for Grp/DChk1 using RNA interference enter mitosis in the presence of impaired DNA integrity, resulting in prolonged mitosis and mitotic catastrophe. Grp/DChk1 is phosphorylated in a Mei-41/DATR-dependent manner in response to hydroxyurea and ionizing radiation, indicating that Mei-41/ATR is an upstream component in the Grp/DChk1 DNA replication and DNA-damage-response pathways. The level of Cdc25(Stg) and phosphorylation status of Cdc2 are modulated in a Grp/DChk1-dependent manner in response to hydroxyurea and irradiation, indicating that these cell-cycle regulators are downstream targets of the Grp/DChk1-dependent DNA replication and DNA-damage responses. By contrast, depletion of Dmnk/DChk2 by RNA interference had little effect on checkpoint responses to hydroxyurea and irradiation. We conclude that Grp/DChk1, and not Dmnk/DChk2, is the main effector kinase involved in G(2)/M checkpoint control in Drosophila cells.
Figures
Fig. 1
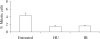
S2 cells display cell-cycle arrest in response to HU or IR. S2 cells were treated with HU for 15 hours or treated with 150 Gy IR followed by 2 hours of recovery. Cells positively stained for PH3 were counted using flow cytometry. S2 cells showed a decrease in the proportion of mitotic cells when treated with HU (1.5%) or IR (1.6%) compared with untreated cells (4.3%). Error bars represent s.e.m. (_n_=13).
Fig. 2
HU- and IR-induced modifications of Cdc25Stg and Cdc2 in control S2 cells but not in Grp/DChk1-depleted S2 cells. Wild-type embryos at 0–2 hours of age (lane 1) expressed the active phosphorylated isoform (form 1) and the unphosphorylated neutral isoform (form 2) of Cdc2. Wild-type embryos at 2–4 hours of age (lane 2) showed accumulation of inhibitory phosphorylated isoforms (forms 3 and 4) of Cdc2. In early embryos (lane 1) Cdc25Stg was expressed, whereas Cdc25Stg protein was almost absent from in older embryos (lane 2). In S2 cells (lanes 3 and 5), Cdc2 isoforms 2–4 were detected and Cdc25Stg was present. In S2 cells treated with 10 mM HU during 15 hours (lane 4) the Cdc2 isoforms 3 and 4 accumulated, Cdc2 isoform 2 was almost absent and Cdc25Stg levels decreased. S2 cells treated with 150 Gy IR followed by 2 hours of recovery (lane 6) showed accumulation of Cdc2 isoform 4 and presence of Cdc2 isoforms 2 and 3. Cdc25Stg levels decreased moderately in response to IR. S2 cells depleted for Grp/DChk1 by RNAi (lanes 7 and 9) expressed Cdc25Stg and all Cdc2 isoforms (forms 2–4). Grp/DChk1-depleted cells treated with 10 mM HU (lane 8) or treated with 150 Gy IR followed by 2 hours recovery (lane 10) did not show modifications of Cdc25Stg and Cdc2. Equal numbers (_n_=20) of Drosophila embryos were loaded in lanes 1 and 2. For lanes 3–10, a background band (Bg) recognized by the Cdc25Stg antibody was used as a loading control.
Fig. 3
Grp/DChk1 is phosphorylated in S2 cells in response to HU or IR. Western-blot analysis of Grp/DChk1 of S2 cells treated with 10 mM HU during 15 hours or with 150 Gy IR followed by 2 hours recovery. Grp/DChk1 was detected as a slower migrating protein in S2 cells treated with HU (A, lane 2) or treated with IR (B, lane 2) compared with untreated S2 cells (A,B, lanes 1,3). A shift in mobility of Grp/DChk1 was not detected in S2 cells treated with HU or IR and calf-intestine alkaline phosphatase (CIP; A,B, lane 4). In both A and B, CP190 protein levels were detected as a loading control.
Fig. 4
(A) Downregulation of Grp/DChk1 protein levels in S2 cells using RNAi. Grp/DChk1 expression in S2 cells (Fig. 4A, lane 1) and S2 cells treated with dsRNA of Grp/DChk1 (Fig. 4A, lane 2). Incubation of S2 cells with dsRNA of Grp/DChk1 for 96 hours reduced Grp/Dchk1 expression to below detection levels. As a loading control, protein levels of γ-tubulin were detected. (B) Grp/DChk1-depleted cells are defective in accomplishing cell-cycle arrest in response to HU or IR. Grp/DChk1 levels were downregulated by RNAi before cells were treated with 10 mM HU for 15 hours or treated with 150 Gy ionizing radiation followed by 2 hours of recovery. Grp/DChk1-depleted cells positively stained for PH3 were counted using flow cytometry. In Grp/DChk1-depleted cells left untreated 5.1% of the cells was in mitosis. In Grp/DChk1-depleted cells treated with HU also 5.1% of the cells was in mitosis and, in IR-treated Grp/DChk1-depleted cells, 4.4% of the cells were in mitosis. Error bars represent s.e.m. (_n_=13).
Fig. 5
Phosphorylation of Grp/DChk1 in response to HU or IR is dependent on the expression of Mei-41/DATR. Western-blot analysis of Grp/DChk1 in control S2 cells and in Mei-41/DATR-depleted S2 cells treated with 10 mM HU during 15 hours or treated with 150 Gy IR followed by 2 hours of recovery. Compared with untreated cells (lanes 1,3,5,7), phosphorylated Grp/DChk1 was detected as a slower-migrating protein in control S2 cells treated with HU (lane 2) or IR (lane 4) and in Mei-41/ATR-depleted S2 cells treated with HU (lane 6) or IR (lane 8), no phosphorylation of Grp/DChk1 was observed. As a loading control, CP190 protein levels were detected.
Fig. 6
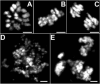
Analysis of mitotic chromosomes from wild-type S2 cells and Grp/DChk1-depleted S2 cells in response to HU or IR. S2 cells were grown on cover slips and mitotic chromosomes were labelled with the anti-PH3 antibody. Images were made by confocal microscopy. (A–C) Mitotic chromosomes of Grp/DChk1-depleted S2 cells under control conditions in prophase (A), metaphase (B) and anaphase (C). (D,E) Fragmentation of mitotic chromosomes in Grp/DChk1-depleted cells treated with 10 mM HU (15 hours) (D) or IR (150 Gy, 2 hours recovery) (E). Scale bar, 1 μm.
Fig. 7
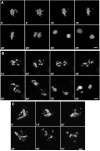
Grp/DChk1-depleted cells display mitotic catastrophe in the presence of incompletely replicated and damaged DNA. (A) Time-lapse confocal analysis allowed normal cell-cycle progression of S2 cells transiently expressing histone H2B-GFP under control conditions. (B,C) Time-lapse confocal analysis of Grp/DChk1-depleted cells transiently expressing histone H2B-GFP and treated with 10 mM HU (B) for 15 hours or with 150 Gy (C) IR followed by 2 hours of recovery. Mitotic chromosomes were irregularly condensed and did not align in the metaphase plate. After prolonged mitosis, chromosomes separated disproportionally and micronuclei were formed. Times are indicated in minutes. Scale bars, 1 μm.
Fig. 8
(A) No mobility shift is observed for Dmnk/DChk2 in response to HU or IR, irrespective of the presence of Grp/DChk1. Western-blot analysis of Dmnk/DChk2 protein in S2 cells and Grp/DChk1-depleted S2 cells treated with 10 mM HU for 15 hours or treated with 150 Gy IR followed by 2 hours of recovery. In S2 cells, Dmnk/DChk2 protein was detected (lane 1) and, in response to HU (lane 2) or IR (lane 3), no shift in mobility of Dmnk/DChk2 was observed. (lane 4) Downregulation of Dmnk/DChk2 protein levels using RNAi. Depletion of Grp/DChk1 using RNAi did not affect the mobility of Dmnk/DChk2 protein in untreated (lanes 5,7), HU-treated (lane 6) or IR-treated (lane 8) S2 cells. As a loading control, γ-tubulin protein levels were detected. (B) Phosphorylation of Grp/DChk1 in response to HU or IR is unaffected in Dmnk/DChk2-depleted cells. Western-blot analysis of Grp/DChk1 in S2 cells and Dmnk/DChk2-depleted cells treated with HU (10 mM, 15 hours) or IR (150 Gy, 2 hours recovery). In S2 cells and in Dmnk/DChk2-depleted cells, a slower-migrating form of Grp/DChk1 was detected in response to HU (lanes 2,6) or IR (lanes 4,8). As a loading control, γ-tubulin protein levels were detected.
Similar articles
- Overexpression of the Drosophila ATR homologous checkpoint kinase Mei-41 induces a G2/M checkpoint in Drosophila imaginal tissue.
Bayer FE, Zimmermann M, Preiss A, Nagel AC. Bayer FE, et al. Hereditas. 2018 Sep 6;155:27. doi: 10.1186/s41065-018-0066-4. eCollection 2018. Hereditas. 2018. PMID: 30202398 Free PMC article. - Drosophila Claspin is required for the G2 arrest that is induced by DNA replication stress but not by DNA double-strand breaks.
Lee EM, Trinh TT, Shim HJ, Park SY, Nguyen TT, Kim MJ, Song YH. Lee EM, et al. DNA Repair (Amst). 2012 Sep 1;11(9):741-52. doi: 10.1016/j.dnarep.2012.06.007. Epub 2012 Jul 15. DNA Repair (Amst). 2012. PMID: 22796626 - Chk2 is required for optimal mitotic delay in response to irradiation-induced DNA damage incurred in G2 phase.
Rainey MD, Black EJ, Zachos G, Gillespie DA. Rainey MD, et al. Oncogene. 2008 Feb 7;27(7):896-906. doi: 10.1038/sj.onc.1210702. Epub 2007 Aug 6. Oncogene. 2008. PMID: 17684483 - Regulation of the G2/M transition by p53.
Taylor WR, Stark GR. Taylor WR, et al. Oncogene. 2001 Apr 5;20(15):1803-15. doi: 10.1038/sj.onc.1204252. Oncogene. 2001. PMID: 11313928 Review. - The multiple checkpoint functions of CHK1 and CHK2 in maintenance of genome stability.
Chen Y, Poon RY. Chen Y, et al. Front Biosci. 2008 May 1;13:5016-29. doi: 10.2741/3060. Front Biosci. 2008. PMID: 18508566 Review.
Cited by
- Drosophila dCBP is involved in establishing the DNA replication checkpoint.
Smolik S, Jones K. Smolik S, et al. Mol Cell Biol. 2007 Jan;27(1):135-46. doi: 10.1128/MCB.01283-06. Epub 2006 Oct 16. Mol Cell Biol. 2007. PMID: 17043110 Free PMC article. - A genome-wide RNAi screen identifies core components of the G₂-M DNA damage checkpoint.
Kondo S, Perrimon N. Kondo S, et al. Sci Signal. 2011 Jan 4;4(154):rs1. doi: 10.1126/scisignal.2001350. Sci Signal. 2011. PMID: 21205937 Free PMC article. - Identification of ER proteins involved in the functional organisation of the early secretory pathway in Drosophila cells by a targeted RNAi screen.
Kondylis V, Tang Y, Fuchs F, Boutros M, Rabouille C. Kondylis V, et al. PLoS One. 2011 Feb 23;6(2):e17173. doi: 10.1371/journal.pone.0017173. PLoS One. 2011. PMID: 21383842 Free PMC article. - Overexpression of the Drosophila ATR homologous checkpoint kinase Mei-41 induces a G2/M checkpoint in Drosophila imaginal tissue.
Bayer FE, Zimmermann M, Preiss A, Nagel AC. Bayer FE, et al. Hereditas. 2018 Sep 6;155:27. doi: 10.1186/s41065-018-0066-4. eCollection 2018. Hereditas. 2018. PMID: 30202398 Free PMC article. - ATM/Chk2 and ATR/Chk1 Pathways Respond to DNA Damage Induced by Movento® 240SC and Envidor® 240SC Keto-Enol Insecticides in the Germarium of Drosophila melanogaster.
González-Marín B, Calderón-Segura ME, Sekelsky J. González-Marín B, et al. Toxics. 2023 Sep 6;11(9):754. doi: 10.3390/toxics11090754. Toxics. 2023. PMID: 37755764 Free PMC article.
References
- Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. - PubMed
- Boddy MN, Russell P. DNA replication checkpoint control. Front. Biosci. 1999;4:D841–D848. - PubMed
- Boddy MN, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. - PubMed
- Brauchle M, Baumer K, Gonczy P. Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr. Biol. 2003;13:819–827. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM66441/GM/NIGMS NIH HHS/United States
- R01 GM066441-02/GM/NIGMS NIH HHS/United States
- R01 GM066441-03/GM/NIGMS NIH HHS/United States
- R01 GM066441-04/GM/NIGMS NIH HHS/United States
- R01 GM066441-01S1/GM/NIGMS NIH HHS/United States
- R01 GM066441/GM/NIGMS NIH HHS/United States
- R01 GM066441-03S1/GM/NIGMS NIH HHS/United States
- R01 GM066441-01/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous