Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide - PubMed (original) (raw)
Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide
Paola Llinas et al. EMBO J. 2005.
Abstract
We report the crystal structure of a soluble form of human urokinase-type plasminogen activator receptor (uPAR/CD87), which is expressed at the invasive areas of the tumor-stromal microenvironment in many human cancers. The structure was solved at 2.7 A in association with a competitive peptide inhibitor of the urokinase-type plasminogen activator (uPA)-uPAR interaction. uPAR is composed of three consecutive three-finger domains organized in an almost circular manner, which generates both a deep internal cavity where the peptide binds in a helical conformation, and a large external surface. This knowledge combined with the discovery of a convergent binding motif shared by the antagonist peptide and uPA allowed us to build a model of the human uPA-uPAR complex. This model reveals that the receptor-binding module of uPA engages the uPAR central cavity, thus leaving the external receptor surface accessible for other protein interactions (vitronectin and integrins). By this unique structural assembly, uPAR can orchestrate the fine interplay with the partners that are required to guide uPA-focalized proteolysis on the cell surface and control cell adhesion and migration.
Figures
Figure 1
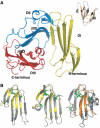
The structure of human uPAR. (A)The overall modular structure of uPAR as a ribbon diagram. The individual uPAR domains are assembled in a right-handed orientation and are coloured yellow (DI), blue (DII) and red (DIII). The inset illustrates the typical β-strand nomenclature (Low et al, 1976) for a three-fingered fold, using bucandin as a model (Torres et al, 2001). Adopting this nomenclature, the 17 β-strands of uPAR encompass the regions 2–6 (βIA), 12–16 (βIB), 23–33 (βIC), 36–46 (βID), 53–57 (βIE) and 64–71 (βIF) in DI; 94–96 (βIIA), 112–114 (βIIB), 121–129 (βIIC), 142–149 (βIID), 155–160 (βIIE) and 164–171 (βIIF) in DII; and 189–199 (βIIIA), 211- 216 (βIIIB), 221–229 (βIIIC), 234–242 (βIIID) and 262–266 (βIIIF) in DIII. The short helical stretches in DIII (αIIIE) encompass residues 244–248 and 250–255. The bended β-strands in DII and DIII are indicated separately. (B) From left to right, superimpositions of bucandin (yellow) on uPAR DI and DII (grey) and of CD59 (orange) on uPAR DIII (grey). Disulphide bonds are coloured green in the superimpositions. For sake of clarity, the positions of the N- and C-termini are indicated in panel A.
Figure 2
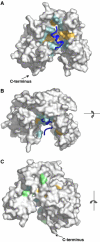
Molecular surface representation of the uPAR–peptide complex. The surface of uPAR is shown in grey, whereas the bound antagonist peptide (AE147) is shown as a ribbon diagram in dark blue. Receptor residues interacting with AE147 are coloured orange (hydrophobic) or cyan (polar). (A) The front side, (B) the upper side and (C) the rear side of the complex, with asparagine-linked glycosylation sites coloured light green. The C-terminal end is shown in panels A and C. In panel B, it is located behind the molecule and hence it cannot be seen by the reader.
Figure 3
Interacting residues in both the antagonist peptide AE147 and uPAR as observed in the crystal structure of the uPAR–peptide complex. The amino-acid sequence of AE147 is shown in the single-letter code with capitals denoting
L
-amino acids and lower case
D
-amino acids. Cha:
L
-β-cyclohexyl-alanine. Residues in black are from human uPAR (Roldan et al, 1990). The corresponding residues from murine uPAR (see text) are coloured blue (Kristensen et al, 1991). Bold letters in blue highlight nonconservative substitutions among these residues in the cavity of human and murine uPAR.
Figure 4
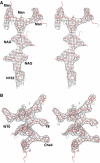
Stereo view of two 2_F_o−_F_c omit maps. (A) A 1.5σ level contoured map for the carbohydrate linked to N162 in uPAR domain DII. (B) A similar contoured map for the bound AE147 (in grey). The refined atomic model of AE147 is shown in stick representation (red). Cha:
L
-β-cyclohexyl-alanine; NAG: _N_-acetylglucosamine; Man: mannose.
Figure 5
Binding of the antagonist peptide in uPAR cavity. (A) The interface between uPAR (illustrated as surface representation) and AE147 (illustrated by a combined ribbon and stick representation in dark blue) is shown from the front side. The interface provided by uPAR is coloured pale orange (hydrophobic) or cyan (polar), whereas the corresponding side chains in AE147 are coloured red. Note that the indole side chain of W10 efficiently engages the deep and prominent hydrophobic hole of the central cavity of uPAR and the side chain of L9 is located proximate to its entrance. (B) Superposition of all eight peptide molecules observed in the unit cell. The figure shows superposition of their Cα backbones and the side chains of Cha4, F5, s6, Y8, L9 and W10 which are involved in binding to uPAR (see text).
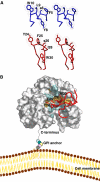
Figure 6
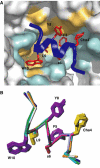
Model of the uPA–uPAR complex. (A) Stereo views of the key residues by which AE147 (blue) and the β-hairpin in GFD of uPA (red) bind to uPAR in our crystal structure and in a GFD–uPAR model, respectively. Note that W10 and L9 of AE147 exhibit a spatial superimposition with Y24 and F25 in uPA. (B) Model for the structure of the human GFD–uPAR complex. The uPAR molecule is shown as a molecular surface representation with the residues interacting with GFD in the same colour code as those used in Figures 2 and 4. The receptor-binding module of uPA is shown in red as a ribbon diagram with the side chains of Y24, F25, I28 and W30, which are highlighted by sticks. A schematic representation of the glycolipid (GPI) anchor is shown to connect the C-terminus of uPAR to a model of the cell membrane. Note that the GPI anchor is not shown to scale and the relative orientation of uPAR to the cell membrane is arbitrarily presented.
Similar articles
- Ligand binding regions in the receptor for urokinase-type plasminogen activator.
Liang OD, Chavakis T, Kanse SM, Preissner KT. Liang OD, et al. J Biol Chem. 2001 Aug 3;276(31):28946-53. doi: 10.1074/jbc.m011347200. J Biol Chem. 2001. PMID: 11501527 - A competitive chromogenic assay to study the functional interaction of urokinase-type plasminogen activator with its receptor.
Rettenberger P, Wilhelm O, Oi H, Weidle UH, Goretzki L, Koppitz M, Lottspeich F, König B, Pessara U, Kramer MD, et al. Rettenberger P, et al. Biol Chem Hoppe Seyler. 1995 Oct;376(10):587-94. doi: 10.1515/bchm3.1995.376.10.587. Biol Chem Hoppe Seyler. 1995. PMID: 8590627 - Cyclo19,31[D-Cys19]-uPA19-31 is a potent competitive antagonist of the interaction of urokinase-type plasminogen activator with its receptor (CD87).
Magdolen V, Bürgle M, de Prada NA, Schmiedeberg N, Riemer C, Schroeck F, Kellermann J, Degitz K, Wilhelm OG, Schmitt M, Kessler H. Magdolen V, et al. Biol Chem. 2001 Aug;382(8):1197-205. doi: 10.1515/BC.2001.150. Biol Chem. 2001. PMID: 11592401 - Structural analysis of the interaction between urokinase-type plasminogen activator and its receptor: a potential target for anti-invasive cancer therapy.
Ploug M, Gårdsvoll H, Jørgensen TJ, Lønborg Hansen L, Danø K. Ploug M, et al. Biochem Soc Trans. 2002 Apr;30(2):177-83. Biochem Soc Trans. 2002. PMID: 12023847 Review. - Structure-function relationships in the interaction between the urokinase-type plasminogen activator and its receptor.
Ploug M. Ploug M. Curr Pharm Des. 2003;9(19):1499-528. doi: 10.2174/1381612033454630. Curr Pharm Des. 2003. PMID: 12871065 Review.
Cited by
- Electrophilic proximity-inducing synthetic adapters enhance universal T cell function by covalently enforcing immune receptor signaling.
Serniuck NJ, Kapcan E, Moogk D, Moore AE, Lake BPM, Denisova G, Hammill JA, Bramson JL, Rullo AF. Serniuck NJ, et al. Mol Ther Oncol. 2024 Jun 24;32(3):200842. doi: 10.1016/j.omton.2024.200842. eCollection 2024 Sep 19. Mol Ther Oncol. 2024. PMID: 39045028 Free PMC article. - Human antibody VH domains targeting uPAR as candidate therapeutics for cancers.
Chu X, Li W, Hines MG, Lyakhov I, Mellors JW, Dimitrov DS. Chu X, et al. Front Oncol. 2023 Oct 9;13:1194972. doi: 10.3389/fonc.2023.1194972. eCollection 2023. Front Oncol. 2023. PMID: 37876962 Free PMC article. - Targeted imaging of uPAR expression in vivo with cyclic AE105 variants.
Leth JM, Newcombe EA, Grønnemose AL, Jørgensen JT, Qvist K, Clausen AS, Knudsen LBS, Kjaer A, Kragelund BB, Jørgensen TJD, Ploug M. Leth JM, et al. Sci Rep. 2023 Oct 11;13(1):17248. doi: 10.1038/s41598-023-43934-w. Sci Rep. 2023. PMID: 37821532 Free PMC article. - Chloroacetamide fragment library screening identifies new scaffolds for covalent inhibition of the TEAD·YAP1 interaction.
Bum-Erdene K, Ghozayel MK, Zhang MJ, Gonzalez-Gutierrez G, Meroueh SO. Bum-Erdene K, et al. RSC Med Chem. 2023 Aug 3;14(9):1803-1816. doi: 10.1039/d3md00264k. eCollection 2023 Sep 19. RSC Med Chem. 2023. PMID: 37731696 Free PMC article. - Circulating Soluble Urokinase Plasminogen Activator Receptor as a Predictive Indicator for COVID-19-Associated Acute Kidney Injury and Mortality: Clinical and Bioinformatics Analysis.
Abdellatif HAA, Sultan BO, Nassar HM, Gomaa MEE, Sakr MG, Riad E, Al-Harbi AI, Abdulhakim JA, Fawzy MS, Abd El-Fadeal NM. Abdellatif HAA, et al. Int J Mol Sci. 2023 Apr 13;24(8):7177. doi: 10.3390/ijms24087177. Int J Mol Sci. 2023. PMID: 37108340 Free PMC article.
References
- Appella E, Robinson EA, Ullrich SJ, Stoppelli MP, Corti A, Cassani G, Blasi F (1987) The receptor-binding sequence of urokinase. A biological function for the growth-factor module of proteases. J Biol Chem 262: 4437–4440 - PubMed
- Bdeir K, Kuo A, Mazar A, Sachais BS, Xiao W, Gawlak S, Harris S, Higazi AA, Cines DB (2000) A region in domain II of the urokinase receptor required for urokinase binding. J Biol Chem 275: 28532–28538 - PubMed
- Blasi F, Carmeliet P (2002) uPAR: a versatile signalling orchestrator. Nat Rev Mol Cell Biol 3: 932–943 - PubMed
- Brooks BR, Karplus M, Petit BM (1988) Proteins: A Theoretical Perspective of Dynamics, Structure and Thermodynamics. New York: Wiley
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous