The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity - PubMed (original) (raw)
The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity
Thomas Clouaire et al. Proc Natl Acad Sci U S A. 2005.
Abstract
We have recently described an evolutionarily conserved protein motif, designated the THAP domain, which defines a previously uncharacterized family of cellular factors (THAP proteins). The THAP domain exhibits similarities to the site-specific DNA-binding domain of Drosophila P element transposase, including a putative metal-coordinating C2CH signature (CX(2-4)CX(35-53)CX(2)H). In this article, we report a comprehensive list of approximately 100 distinct THAP proteins in model animal organisms, including human nuclear proapoptotic factors THAP1 and DAP4/THAP0, transcriptional repressor THAP7, zebrafish orthologue of cell cycle regulator E2F6, and Caenorhabditis elegans chromatin-associated protein HIM-17 and cell-cycle regulators LIN-36 and LIN-15B. In addition, we demonstrate the biochemical function of the THAP domain as a zinc-dependent sequence-specific DNA-binding domain belonging to the zinc-finger superfamily. In vitro binding-site selection allowed us to identify an 11-nucleotide consensus DNA-binding sequence specifically recognized by the THAP domain of human THAP1. Mutations of single nucleotide positions in this sequence abrogated THAP-domain binding. Experiments with the zinc chelator 1,10-o-phenanthroline revealed that the THAP domain is a zinc-dependent DNA-binding domain. Site-directed mutagenesis of single cysteine or histidine residues supported a role for the C2CH motif in zinc coordination and DNA-binding activity. The four other conserved residues (P, W, F, and P), which define the THAP consensus sequence, were also found to be required for DNA binding. Together with previous genetic data obtained in C. elegans, our results suggest that cellular THAP proteins may function as zinc-dependent sequence-specific DNA-binding factors with roles in proliferation, apoptosis, cell cycle, chromosome segregation, chromatin modification, and transcriptional regulation.
Figures
Fig. 1.
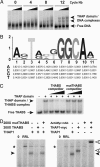
THAP proteins in model animal organisms. (A) Identification of a consensus THAP domain in the zebrafish orthologue of cell-cycle transcription factor E2F6. Shown is
clustalw
multiple alignment of zebrafish E2F6 with human E2F6 and the THAP domain of human THAP1. Conserved residues are boxed. Black boxes indicate identical residues, whereas boxes shaded in gray show similar amino acids. The consensus THAP motif, defined by the C2CH signature and four other invariant residues (P26, W36, F58, and P78 in human THAP1), is shown above the alignment. (B) Primary structures of C. elegans THAP proteins with known functions. THAP domains are shown in black. Divergent THAP domains containing the C2CH signature are shown in gray. Known protein motifs are indicated. The transcriptional-corepressor function of CtBP has not yet been confirmed in C. elegans.
Fig. 2.
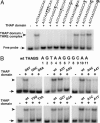
The THAP domain is a sequence-specific DNA-binding domain. (A) Identification of THAP-domain DNA-target sequence by SELEX. DNA recovered after 0, 4, 8, and 12 rounds of selection was labeled and incubated with increasing amounts of recombinant THAP domain (1, 6, and 60 ng, respectively). Resulting protein–DNA complexes were analyzed in EMSA. (B) Identification of a consensus THABS. The oligonucleotide sequences recovered after 12 rounds of selection were analyzed by using the motif-discovery program
meme
. The position-specific probability matrix returned by
meme
is given. (C) Specificity of THAP-domain–THABS interaction. Recombinant THAP domain was incubated with THABS probe in the absence or presence of increasing amounts (50-, 150-, and 250-fold molar excess) of either THABS (AGTAAGGGCAA) or mutTHABS (AGTAA
TTT
CAA) unlabeled competitor. The protein–DNA complexes were analyzed in EMSA. (D) Binding of full-length THAP1 to the THABS. In vitro translated THAP1 or THAP1-Myc was incubated with labeled THABS probe in the absence or presence of a 200-fold molar excess of either THABS or mutTHABS unlabeled competitor, and protein–DNA complexes were analyzed in EMSA. For the supershift experiment, THAP1-Myc was incubated with the THABS probe in the presence of anti-Myc mAb. RRL, THABS probe incubated with unprogrammed RRL; black arrowhead, THAP1–THABS DNA complex; white arrowhead, THAP1-Myc–THABS DNA complex; *, nonspecific complex.
Fig. 3.
Mutations of single nucleotide positions in the THABS abrogate recognition by the THAP domain of human THAP1. (A) The GGCA core motif of the THABS is required for THAP-domain–THABS interaction. EMSAs were performed by using recombinant THAP domain and labeled oligonucleotides bearing mutations in the GGCA core motif of the THABS. (B) Scanning mutagenesis of the THABS sequence reveals that bases upstream of the GGCA core motif modulate the strength and affinity of THAP-domain–THABS interaction. EMSAs were performed by using recombinant THAP-domain and DNA targets bearing single-point mutations in the THABS sequence.
Fig. 4.
The THAP domain is a zinc-dependent DNA-binding domain. (A) Inhibition of THAP-domain DNA-binding activity by metal-chelating agents EDTA and 1,10-_o_-phenanthroline. THAP-domain–THABS DNA complexes were analyzed by EMSA. Lane 0, THABS probe alone; UT, THABS probe incubated with untreated THAP domain; MeOH, methanol vehicle alone. (B) Role of zinc in THAP-domain DNA-binding activity. Recombinant THAP domain was treated with 5 mM 1,10-_o_-phenanthroline, and the chloride of Zn2+, Ca2+, Mg2+, or Fe2+ was subsequently added to the binding reactions before analysis of THAP-domain–DNA complexes (arrowhead) by EMSA.
Fig. 5.
The C2CH signature and the four other invariant residues of the THAP domain are essential for DNA binding. (A) Generation of THAP1 mutants in the consensus THAP motif. Wild-type THAP1 (wt) and single-point mutants THAP1-C5A, THAP1-C10A, THAP1-C54A, THAP1-H57A, THAP1-P26A, THAP1-W36A, THAP1-F58A, and THAP1-P78A were translated in vitro in RRL in the presence of 35S-labeled methionine and analyzed by SDS/PAGE and autoradiography. Molecular mass markers are shown on the left (kDa). (B) Mutation of single cysteine or histidine residues of the C2CH signature abrogates THAP-domain DNA-binding activity. EMSAs were performed with the THABS probe and THAP1 wild-type (wt) or mutant proteins (C5A, C10A, C54A, and H57A). For comparison, wild-type THAP1 was incubated with 5 mM 1,10-_o_-phenanthroline or methanol vehicle alone (MeOH). RRL, unprogrammed RRL; arrowhead, THAP1–THABS DNA complex; *, nonspecific complexes. (C) Mutation of the four other conserved residues of the THAP domain (P, W, F, and P) abrogates DNA binding. EMSAs were performed with the THABS probe and THAP1 wild-type (wt) or mutant proteins (P26A, W36A, F58A, and P78A).
Similar articles
- Structure-function analysis of the THAP zinc finger of THAP1, a large C2CH DNA-binding module linked to Rb/E2F pathways.
Bessière D, Lacroix C, Campagne S, Ecochard V, Guillet V, Mourey L, Lopez F, Czaplicki J, Demange P, Milon A, Girard JP, Gervais V. Bessière D, et al. J Biol Chem. 2008 Feb 15;283(7):4352-63. doi: 10.1074/jbc.M707537200. Epub 2007 Dec 11. J Biol Chem. 2008. PMID: 18073205 - Solution structure of the THAP domain from Caenorhabditis elegans C-terminal binding protein (CtBP).
Liew CK, Crossley M, Mackay JP, Nicholas HR. Liew CK, et al. J Mol Biol. 2007 Feb 16;366(2):382-90. doi: 10.1016/j.jmb.2006.11.058. Epub 2006 Nov 18. J Mol Biol. 2007. PMID: 17174978 - The THAP-zinc finger protein THAP1 associates with coactivator HCF-1 and O-GlcNAc transferase: a link between DYT6 and DYT3 dystonias.
Mazars R, Gonzalez-de-Peredo A, Cayrol C, Lavigne AC, Vogel JL, Ortega N, Lacroix C, Gautier V, Huet G, Ray A, Monsarrat B, Kristie TM, Girard JP. Mazars R, et al. J Biol Chem. 2010 Apr 30;285(18):13364-71. doi: 10.1074/jbc.M109.072579. Epub 2010 Mar 3. J Biol Chem. 2010. PMID: 20200153 Free PMC article. - NMR studies of a new family of DNA binding proteins: the THAP proteins.
Gervais V, Campagne S, Durand J, Muller I, Milon A. Gervais V, et al. J Biomol NMR. 2013 May;56(1):3-15. doi: 10.1007/s10858-012-9699-1. Epub 2013 Jan 11. J Biomol NMR. 2013. PMID: 23306615 Review. - DYT6 dystonia: review of the literature and creation of the UMD Locus-Specific Database (LSDB) for mutations in the THAP1 gene.
Blanchard A, Ea V, Roubertie A, Martin M, Coquart C, Claustres M, Béroud C, Collod-Béroud G. Blanchard A, et al. Hum Mutat. 2011 Nov;32(11):1213-24. doi: 10.1002/humu.21564. Epub 2011 Sep 15. Hum Mutat. 2011. PMID: 21793105 Review.
Cited by
- The dystonia gene THAP1 controls DNA double-strand break repair choice.
Shinoda K, Zong D, Callen E, Wu W, Dumitrache LC, Belinky F, Chari R, Wong N, Ishikawa M, Stanlie A, Multhaupt-Buell T, Sharma N, Ozelius L, Ehrlich M, McKinnon PJ, Nussenzweig A. Shinoda K, et al. Mol Cell. 2021 Jun 17;81(12):2611-2624.e10. doi: 10.1016/j.molcel.2021.03.034. Epub 2021 Apr 14. Mol Cell. 2021. PMID: 33857404 Free PMC article. - Genetic innovation in vertebrates: gypsy integrase genes and other genes derived from transposable elements.
Chalopin D, Galiana D, Volff JN. Chalopin D, et al. Int J Evol Biol. 2012;2012:724519. doi: 10.1155/2012/724519. Epub 2012 Aug 13. Int J Evol Biol. 2012. PMID: 22928150 Free PMC article. - The genetics of dystonia: new twists in an old tale.
Charlesworth G, Bhatia KP, Wood NW. Charlesworth G, et al. Brain. 2013 Jul;136(Pt 7):2017-37. doi: 10.1093/brain/awt138. Epub 2013 Jun 17. Brain. 2013. PMID: 23775978 Free PMC article. Review. - THAP and ATF-2 regulated sterol carrier protein-2 promoter activities in the larval midgut of the yellow fever mosquito, Aedes aegypti.
Peng R, Fu Q, Hong H, Schwaegler T, Lan Q. Peng R, et al. PLoS One. 2012;7(10):e46948. doi: 10.1371/journal.pone.0046948. Epub 2012 Oct 4. PLoS One. 2012. PMID: 23056538 Free PMC article. - DNA-binding and -bending activities of SAP30L and SAP30 are mediated by a zinc-dependent module and monophosphoinositides.
Viiri KM, Jänis J, Siggers T, Heinonen TY, Valjakka J, Bulyk ML, Mäki M, Lohi O. Viiri KM, et al. Mol Cell Biol. 2009 Jan;29(2):342-56. doi: 10.1128/MCB.01213-08. Epub 2008 Nov 17. Mol Cell Biol. 2009. PMID: 19015240 Free PMC article.
References
- Roussigne, M., Kossida, S., Lavigne, A. C., Clouaire, T., Ecochard, V., Glories, A., Amalric, F. & Girard, J. P. (2003) Trends Biochem. Sci. 28, 66–69. - PubMed
- Roussigne, M., Cayrol, C., Clouaire, T., Amalric, F. & Girard, J. P. (2003) Oncogene 22, 2432–2442. - PubMed
- Deiss, L. P., Feinstein, E., Berissi, H., Cohen, O. & Kimchi, A. (1995) Genes Dev. 9, 15–30. - PubMed
- Lin, Y., Khokhlatchev, A., Figeys, D. & Avruch, J. (2002) J. Biol. Chem. 277, 47991–48001. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases