Genomewide production of multipurpose alleles for the functional analysis of the mouse genome - PubMed (original) (raw)
Comparative Study
. 2005 May 17;102(20):7221-6.
doi: 10.1073/pnas.0502273102. Epub 2005 May 3.
Silke De-Zolt, Petra Van Sloun, Melanie Hollatz, Thomas Floss, Jens Hansen, Joachim Altschmied, Claudia Seisenberger, Norbert B Ghyselinck, Patricia Ruiz, Pierre Chambon, Wolfgang Wurst, Harald von Melchner
Affiliations
- PMID: 15870191
- PMCID: PMC1129123
- DOI: 10.1073/pnas.0502273102
Comparative Study
Genomewide production of multipurpose alleles for the functional analysis of the mouse genome
Frank Schnütgen et al. Proc Natl Acad Sci U S A. 2005.
Abstract
A type of retroviral gene trap vectors has been developed that can induce conditional mutations in most genes expressed in mouse embryonic stem (ES) cells. The vectors rely on directional site-specific recombination systems that can repair and re-induce gene trap mutations when activated in succession. After the gene traps are inserted into the mouse genome, genetic mutations can be produced at a particular time and place in somatic cells. In addition to their conditional features, the vectors create multipurpose alleles amenable to a wide range of post-insertional modifications. Here we have used these directional recombination vectors to assemble the largest library of ES cell lines with conditional mutations in single genes yet assembled, presently totaling 1,000 unique genes. The trapped ES cell lines, which can be ordered from the German Gene Trap Consortium, are freely available to the scientific community.
Figures
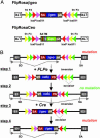
Fig. 1.
Conditional gene trap vectors and mechanism of gene inactivation. (A) Schematic representation of the retroviral gene trap vectors. LTR, long terminal repeat; frt (yellow triangles) and F3 (green triangles), heterotypic target sequences for the FLPe recombinase; loxP (red triangles) and lox511 (purple triangles), heterotypic target sequences for the Cre-recombinase; SA, splice acceptor; βgeo, β-galactosidase/neomycin phosphotransferase fusion gene; pA, bovine growth hormone polyadenylation sequence; TM, human CD2 receptor transmembrane domain. (B) Conditional gene inactivation by a SAβgeopA cassette. The SAβgeopA cassette flanked by recombinase target sites (RTs) in a FlEx configuration is illustrated after integration into an intron of an expressed gene. Transcripts (shown as gray arrows) initiated at the endogenous promoter are spliced from the splice donor (SD) of an endogenous exon (here, exon 1) to the SA of the SAβgeopA cassette. Thereby the βgeo reporter gene is expressed and the endogenous transcript is captured and prematurely terminated at the cassette's pA causing a mutation. In step 1, FLPe inverts the SAβgeopA cassette onto the antisense, noncoding strand at either frt (shown) or F3 (not shown) RTs and positions frt and F3 sites between direct repeats of F3 and frt RTs, respectively. By simultaneously excising the heterotypic RTs (step 2), the cassette is locked against reinversion because the remaining frt and F3 RTs cannot recombine. This inversion reactivates normal splicing between the endogenous splice sites, thereby repairing the mutation. Cre-mediated inversion in steps 3 and 4 repositions the SAβgeopA cassette back onto the sense, coding strand and reinduces the mutation. Note that the recombination products of steps 1 and 3 are transient and transformed into the stable products of step 2 and 4, respectively (15).
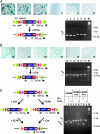
Fig. 2.
Site-specific recombinase-induced inversions in FlipRosaβgeo-trapped ES cell lines. (A and B) ES cells were infected with FlipRosaβgeo virus and selected in G418. X-Gal-positive sublines (blue) were electroporated with FLPe (A) or Cre (B) expression plasmids and stained with X-Gal after incubating for 10 days. DNA extracted from blue and white sublines was subjected to a multiplex PCR to identify inversions. Primer positions within FlipRosaβgeo are indicated by large arrows; allele-specific amplification products are visualized on ethidium bromide-stained gels to the right. (C) Sublines of the FS4B6 ES cell line harboring Cre- or FLPe-inverted gene trap insertions were electroporated with both FLPe- and Cre-expression plasmids. The amplification products obtained from the progeny lines by allele-specific PCR are visualized on the ethidium bromide-stained gel to the right. t, trapped allele; inv, inverted allele; re-inv, reinverted allele; M, molecular weight marker (1 kb + ladder, Invitrogen). FS4B6 (1anes 1-3), parental FlipRosaβgeo-trapped ES cell line; FS4B6 C14 (lanes 4-6), Cre-inverted subline; and FS4B6 F14 (lanes 7-9), FLPe-inverted subline.
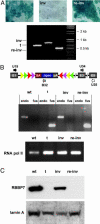
Fig. 3.
Conditional mutation induced by a FlipRosaβgeo gene trap insertion in the RBBP7 gene (ENSEMBL ID: ENSMUSG00000031353). The Q017B06 gene trap cell line (t) was transiently transfected with a FLPe expression plasmid, and several sublines with inverted gene trap cassettes were identified by X-Gal staining and allele-specific PCR (inv). Inverted sublines were then electroporated with a Cre-expression plasmid and enriched for reinversions by selecting in G418 (re-inv). (A) X-Gal staining (Upper) and allele-specific PCR amplification products (Lower) from the trapped RBBP7 locus in trapped (t), inverted (inv), and reinverted (re-inv) Q017B06 cell lines. Primers used for the multiplex PCRs were identical to those shown in the diagrams of Fig. 2. (B) RT-PCR for the amplification of RBBP7 wild-type and trapped fusion transcripts expressed in Q017B06 cells before and after exposure to FLPe and Cre recombinases. The positions of the primers used are shown on top, wherein U19 = 5′-GCT CTT GAC TAG CGA GAG AGA AG-3′, B32 = 5′-CAA GGC GAT TAA GTT GGG TAA CG-3′, U34 = 5′-CCA GAA GGA AAG GAT TAT GC-3′, and U35 = 5′-ACA GAG CAA ATG ACC CAA GG-3′. Amplification products are visualized below on ethidium bromide-stained gels. Amplification of the RNA polymerase II (RNA pol II) transcript serves as a positive control. wt, parental ES cells; t, trapped Q017B06 cells; inv, inverted Q017B06 subline; re-inv, reinverted Q017B06 subline; endo, endogenous transcript; fus, fusion transcript. (C) Western blot analysis of the RBBP7 protein expressed in Q017B06 cells. Crude cell lysates from the F1 (wt), Q017B06 (t), inverted Q017B06 (inv), and reinverted Q017B06 (re-inv) ES cells were resolved by SDS/PAGE and analyzed by Western blotting using the anti-RbAp46 antibody. The anti-lamin A antibody served as a loading control.
Fig. 4.
Conditional mutation induced by a FlipRosaCeo gene trap insertion in the Glt28d1 gene (ENSEMBL ID ENSMUST00000040338). The M117B08 gene trap line was treated with recombinases and processed as described for Q017B06 in the legend to Fig. 3, except that Cre was used for the first inversion and FLPe for the second. (A) Allele-specific PCR of the trapped Glt28d1 locus in trapped (t), inverted (inv), and reinverted (re-inv) cell lines. (B) RT-PCR of Glt28d1wild type and Glt28d1/gene trap fusion transcripts expressed in M117B08 cells before and after exposure to Cre and FLPe recombinases. The positions of the respective primers within the trapped gene are shown on top, wherein M117B8s = 5′-GAG AGT GCT GGC CAG CTG GAA C-3′, G01 = 5′-CAA GTT GAT GTC CTG ACC CAA G-3′, and M117B8as1 = 5′-CCA CCA TAC TCC ACA CAC TCT G-3′. Amplification products are visualized on ethidium bromide-stained gels below. Amplification of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript serves as a positive control. wt, parental ES cells; t, trapped M117B08; inv, inverted M117B08 subline; re-inv, reinverted M117B08 subline; endo, endogenous transcript; fus, fusion transcript. (C) Northern blot analysis of Glt28d1 transcripts expressed in M117B08 cells. Samples (2 μg) of polyadenylated RNAs from wild-type (wt), Q017B06 (t), inverted M117B08 (inv), and reinverted M117B08 (re-inv) ES cells were fractionated on 1% formaldehyde/agarose gels and hybridized to a 32P-labeled Glt28d1 cDNA probe. The Glt28d1 probe was obtained by asymmetric RT-PCR using a reverse primer in exon 10 to amplify sequences upstream of the insertions site. The loading of each lane was then assessed by using a GAPDH probe. endo, endogenous transcript; fus, fusion transcript; Glt28d1, Glt28d1 transcript.
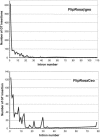
Fig. 5.
Distribution of gene trap insertions according to the position of the trapped intron within genes. The data are based on National Center for Biotechnology Information (NCBI) mouse genome build 33 and RefSeq release 8.
Similar articles
- Generation of multipurpose alleles for the functional analysis of the mouse genome.
Schnütgen F. Schnütgen F. Brief Funct Genomic Proteomic. 2006 Mar;5(1):15-8. doi: 10.1093/bfgp/ell009. Epub 2006 Feb 22. Brief Funct Genomic Proteomic. 2006. PMID: 16769672 Review. - High-throughput trapping of secretory pathway genes in mouse embryonic stem cells.
De-Zolt S, Schnütgen F, Seisenberger C, Hansen J, Hollatz M, Floss T, Ruiz P, Wurst W, von Melchner H. De-Zolt S, et al. Nucleic Acids Res. 2006 Feb 13;34(3):e25. doi: 10.1093/nar/gnj026. Nucleic Acids Res. 2006. PMID: 16478711 Free PMC article. - Gene trap mutagenesis: a functional genomics approach towards reproductive research.
Lee T, Shah C, Xu EY. Lee T, et al. Mol Hum Reprod. 2007 Nov;13(11):771-9. doi: 10.1093/molehr/gam069. Epub 2007 Sep 23. Mol Hum Reprod. 2007. PMID: 17890780 - EUCOMM--the European conditional mouse mutagenesis program.
Friedel RH, Seisenberger C, Kaloff C, Wurst W. Friedel RH, et al. Brief Funct Genomic Proteomic. 2007 Sep;6(3):180-5. doi: 10.1093/bfgp/elm022. Epub 2007 Oct 29. Brief Funct Genomic Proteomic. 2007. PMID: 17967808 - Gene-trap mutagenesis: past, present and beyond.
Stanford WL, Cohn JB, Cordes SP. Stanford WL, et al. Nat Rev Genet. 2001 Oct;2(10):756-68. doi: 10.1038/35093548. Nat Rev Genet. 2001. PMID: 11584292 Review.
Cited by
- Conditional control of gene function by an invertible gene trap in zebrafish.
Ni TT, Lu J, Zhu M, Maddison LA, Boyd KL, Huskey L, Ju B, Hesselson D, Zhong TP, Page-McCaw PS, Stainier DY, Chen W. Ni TT, et al. Proc Natl Acad Sci U S A. 2012 Sep 18;109(38):15389-94. doi: 10.1073/pnas.1206131109. Epub 2012 Aug 20. Proc Natl Acad Sci U S A. 2012. PMID: 22908272 Free PMC article. - The Hole and the Whole: Lessons from Manipulation of Nipbl Deficiency.
Gelb BD. Gelb BD. PLoS Biol. 2016 Sep 8;14(9):e2000494. doi: 10.1371/journal.pbio.2000494. eCollection 2016 Sep. PLoS Biol. 2016. PMID: 27606622 Free PMC article. - Floxin, a resource for genetically engineering mouse ESCs.
Singla V, Hunkapiller J, Santos N, Seol AD, Norman AR, Wakenight P, Skarnes WC, Reiter JF. Singla V, et al. Nat Methods. 2010 Jan;7(1):50-2. doi: 10.1038/nmeth.1406. Epub 2009 Dec 6. Nat Methods. 2010. PMID: 19966808 Free PMC article. - Efficient conditional and promoter-specific in vivo expression of cDNAs of choice by taking advantage of recombinase-mediated cassette exchange using FlEx gene traps.
Schebelle L, Wolf C, Stribl C, Javaheri T, Schnütgen F, Ettinger A, Ivics Z, Hansen J, Ruiz P, von Melchner H, Wurst W, Floss T. Schebelle L, et al. Nucleic Acids Res. 2010 May;38(9):e106. doi: 10.1093/nar/gkq044. Epub 2010 Feb 5. Nucleic Acids Res. 2010. PMID: 20139417 Free PMC article. - FlEx-based transgenic reporter lines for visualization of Cre and Flp activity in live zebrafish.
Boniface EJ, Lu J, Victoroff T, Zhu M, Chen W. Boniface EJ, et al. Genesis. 2009 Jul;47(7):484-91. doi: 10.1002/dvg.20526. Genesis. 2009. PMID: 19415631 Free PMC article.
References
- Cox, R. D. & Brown, S. D. (2003) Curr. Opin. Genet. Dev. 13, 278-283. - PubMed
- Brown, S. D. & Balling, R. (2001) Curr. Opin. Genet. Dev. 11, 268-273. - PubMed
- Floss, T. & Wurst, W. (2002) Methods Mol. Biol. 185, 347-379. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials