ACT-5 is an essential Caenorhabditis elegans actin required for intestinal microvilli formation - PubMed (original) (raw)
ACT-5 is an essential Caenorhabditis elegans actin required for intestinal microvilli formation
A J MacQueen et al. Mol Biol Cell. 2005 Jul.
Abstract
Investigation of Caenorhabditis elegans act-5 gene function revealed that intestinal microvillus formation requires a specific actin isoform. ACT-5 is the most diverged of the five C. elegans actins, sharing only 93% identity with the other four. Green fluorescent protein reporter and immunofluorescence analysis indicated that act-5 gene expression is limited to microvillus-containing cells within the intestine and excretory systems and that ACT-5 is apically localized within intestinal cells. Animals heterozygous for a dominant act-5 mutation looked clear and thin and grew slowly. Animals homozygous for either the dominant act-5 mutation, or a recessive loss of function mutant, exhibited normal morphology and intestinal cell polarity, but died during the first larval stage. Ultrastructural analysis revealed a complete loss of intestinal microvilli in homozygous act-5 mutants. Forced expression of ACT-1 under the control of the act-5 promoter did not rescue the lethality of the act-5 mutant. Together with immuno-electron microscopy experiments that indicated ACT-5 is enriched within microvilli themselves, these results suggest a microvillus-specific function for act-5, and further, they raise the possibility that specific actins may be specialized for building microvilli and related structures.
Figures
Figure 1.
Excretory and alimentary tract cells express act-5. (a and b) GFP expression, controlled by act-5 promoter sequences, is observed within each of the 20 pairs of intestinal cells that form a tube along most of the length of the animal; the H-shaped excretory cell (EC), pharyngeal-intestinal valve cells (VPI), and rectal epithelial cells (REP) also express the transgene reporter. The act-5::gfp transgene has been lost from intestinal cells of the animal in b, allowing better visualization of the EC, REP, and VPI cells. Anterior is left.
Figure 2.
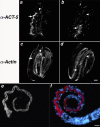
anti-ACT-5 specifically labels the apical intestine. (a–d) Wild-type embryos costained with purified anti-ACT-5 antibodies (a and b) and C4, a pan-reactive anti-actin mAb (c and d). Images represent single confocal optical sections from the middle third (a and c) and top third (b and d) of the same threefold embryo. Arrows indicate the posterior (a and c) and anterior (b and d) of the intestine, where robust anti-ACT-5 label, but very little C4 staining is present (a vs. c); arrowheads point to a single longitudinal body wall muscle quadrant, which is not detected with anti-ACT-5 but readily labeled with C4 (b vs. d). Bar, 5 μm (a–d). (e and f) Wild-type L1 animals stained with anti-ACT-5, in white (e) or red (f); DAPI-stained nuclei are shown in blue (f). ACT-5 label decorates the lumenal surface of the intestine, showing as a tube along most of the length of this L1 animal. Images in e and f are projections of serial image slices that encompass approximately one-half the volume of an intestine and derive from the central portion of the intestine; this processing reveals the unlabeled lumenal area at the center of the intestine. Bar, 10 μm (e and f).
Figure 3.
Actin protein alignment and sequence changes caused by act-5 mutations. C. elegans actin protein alignment. ClustalW 1.83 alignment (Thompson et al., 1994) between the five C. elegans actins (CeACT-1–5, accession nos. CAB04678, CAB04675, CAB04676, AAB04575, and CAB05817, respectively) and the predicted Caenorhabditis briggsae ACT-5 (CbACT-5, accession no. CAE71353). The protein sequence for C. elegans ACT-1 and ACT-3 are identical and denoted as CeACT-1_3. Identical residues are shaded in black, conserved changes in gray. Sequence coordinates are given for the entire alignment, which is offset 1 residue for CeACT-5 and CbACT-5. The boxed region at residues 220–232 indicates the synthetic peptide used to elicit anti-ACT-5 antibodies. Asterisk indicates the position of the premature stop codon in dt2019; down arrows denote the region deleted in dt2017 and replaced by asparagine.
Figure 4.
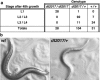
act-5 is essential for growth. (a) Table presents an analysis of the growth of individual animals in a brood, after 2 d. Animals were assayed by PCR to identify genotype after their phenotype was recorded. act-5 (dt2017) homozygotes correspond to animals that fail to grow beyond the first larval stage (L1), even after 2 d of development. (b) Adults carrying one copy of act-5 (dt2017) are skinny and clear (right), compared with wild-type adults (left). Images were taken with bright field optics.
Figure 5.
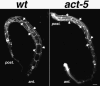
Intestinal cell polarization does not require act-5 function. MH27 staining (white) labels apical regions of polarized cells, giving rise to a web-like pattern along the bodies of the wild-type (left) and act-5(dt2017) mutant (right) worms. Although act-5 mutant intestinal lumens seem slightly deformed relative to control animals, MH27 is properly restricted to apical cell regions in both genotypes. Projections encompass the entire volume of each animal. Anterior (ant.) and posterior (post.) ends of the worm are indicated. Bar, 10 μm.
Figure 6.
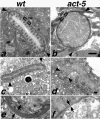
Intestinal microvilli are defective or absent from ACT-5–depleted animals. In wild-type ultrastructural preparations (a and c), intestinal cross sections reveal tightly packed, finger-like projections that correspond to microvilli projecting from the lumenal surface. At the base of these projections is an electron-dense region called the terminal web (TW). A small segment of the terminal web is enclosed by brackets in a and b. Two finger-like projections pointing toward the cytoplasmic side of the intestinal cells are the apical junctions (arrowheads) that mark the boundary between apical regions of two intestinal cells. Microvilli are absent from intestinal cross sections of act-5 (dt2017) animals (b). act-5(RNAi) animals frequently exhibited one shortened apical region (distance between apical junctions) on one of a pair of intestinal cells; this shortened apical surface was typically associated with an abnormally thick terminal web (d). Equivalent higher magnification views of a portion of the thick (e) and normal terminal web width (f) from the micrograph shown in d reveal the difference in thickness as the gap between the arrows (e and f). Control RNAi cross section from an L2–L3 stage animal shown in c. Bar for a and b in b; for c and d in d, 500 nm.
Figure 7.
anti-ACT-5 antisera labels microvilli on thin sections. Gold particles (10 nm), detecting anti-ACT-5 antisera, exhibit robust labeling on microvilli of wild-type intestinal cross sections (a–c), whereas exhibiting little or no label in other areas of the section (muscle cells in f). MH33 antisera, marked by 5-nm gold particles, labels the terminal web and apical junction areas (b and c). C4 (marked by 5-nm gold particles), a pan-reactive monoclonal anti-actin antibody, which detects several, if not all, C. elegans actins, labels microvilli (d) in addition to several additional cells within the worm; muscle cell labeling is shown in e. Insets in b, c, and d show enlarged regions to enable better visualization of 5-nm gold particles. Bar, 500 nm.
Similar articles
- Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans.
Ono K, Parast M, Alberico C, Benian GM, Ono S. Ono K, et al. J Cell Sci. 2003 May 15;116(Pt 10):2073-85. doi: 10.1242/jcs.00421. Epub 2003 Apr 1. J Cell Sci. 2003. PMID: 12679387 - The third and fourth tropomyosin isoforms of Caenorhabditis elegans are expressed in the pharynx and intestines and are essential for development and morphology.
Anyanful A, Sakube Y, Takuwa K, Kagawa H. Anyanful A, et al. J Mol Biol. 2001 Oct 26;313(3):525-37. doi: 10.1006/jmbi.2001.5052. J Mol Biol. 2001. PMID: 11676537 - Conditional dominant mutations in the Caenorhabditis elegans gene act-2 identify cytoplasmic and muscle roles for a redundant actin isoform.
Willis JH, Munro E, Lyczak R, Bowerman B. Willis JH, et al. Mol Biol Cell. 2006 Mar;17(3):1051-64. doi: 10.1091/mbc.e05-09-0886. Epub 2006 Jan 11. Mol Biol Cell. 2006. PMID: 16407404 Free PMC article. - Generation of intestinal surface: an absorbing tale.
Walton KD, Freddo AM, Wang S, Gumucio DL. Walton KD, et al. Development. 2016 Jul 1;143(13):2261-72. doi: 10.1242/dev.135400. Development. 2016. PMID: 27381224 Free PMC article. Review. - F-actin bundles are derivatives of microvilli: What does this tell us about how bundles might form?
DeRosier DJ, Tilney LG. DeRosier DJ, et al. J Cell Biol. 2000 Jan 10;148(1):1-6. J Cell Biol. 2000. PMID: 10629213 Free PMC article. Review. No abstract available.
Cited by
- Small GTPases promote actin coat formation on microsporidian pathogens traversing the apical membrane of Caenorhabditis elegans intestinal cells.
Szumowski SC, Estes KA, Popovich JJ, Botts MR, Sek G, Troemel ER. Szumowski SC, et al. Cell Microbiol. 2016 Jan;18(1):30-45. doi: 10.1111/cmi.12481. Epub 2015 Jul 28. Cell Microbiol. 2016. PMID: 26147591 Free PMC article. - Collagen and actin network mediate antiviral immunity against Orsay virus in C. elegans intestinal cells.
Zhou Y, Chen H, Zhong W, Tao YJ. Zhou Y, et al. PLoS Pathog. 2024 Jan 8;20(1):e1011366. doi: 10.1371/journal.ppat.1011366. eCollection 2024 Jan. PLoS Pathog. 2024. PMID: 38190406 Free PMC article. - Transcriptional adaptation in Caenorhabditis elegans.
Serobyan V, Kontarakis Z, El-Brolosy MA, Welker JM, Tolstenkov O, Saadeldein AM, Retzer N, Gottschalk A, Wehman AM, Stainier DY. Serobyan V, et al. Elife. 2020 Jan 17;9:e50014. doi: 10.7554/eLife.50014. Elife. 2020. PMID: 31951195 Free PMC article. - Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety.
You YJ, Kim J, Raizen DM, Avery L. You YJ, et al. Cell Metab. 2008 Mar;7(3):249-57. doi: 10.1016/j.cmet.2008.01.005. Cell Metab. 2008. PMID: 18316030 Free PMC article. - Mutation of the Enterohemorrhagic Escherichia coli Core LPS Biosynthesis Enzyme RfaD Confers Hypersusceptibility to Host Intestinal Innate Immunity In vivo.
Kuo CJ, Chen JW, Chiu HC, Teng CH, Hsu TI, Lu PJ, Syu WJ, Wang ST, Chou TC, Chen CS. Kuo CJ, et al. Front Cell Infect Microbiol. 2016 Aug 12;6:82. doi: 10.3389/fcimb.2016.00082. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27570746 Free PMC article.
References
- Bossinger, O., Fukushige, T., Claeys, M., Borgonie, G., and McGhee, J. D. (2004). The apical disposition of the Caenorhabditis elegans intestinal terminal web is maintained by LET-413. Dev. Biol. 268, 448–456. - PubMed
- Buechner, M. (2002). Tubes and the single C. elegans excretory cell. Trends Cell Biol. 12, 479–484. - PubMed
- Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W., and Prasher, D. C. (1994). Green fluorescent protein as a marker for gene expression. Science 263, 802–805. - PubMed
- Croce, A., Cassata, G., Disanza, A., Gagliani, M. C., Tacchetti, C., Malabarba, M. G., Carlier, M. F., Scita, G., Baumeister, R., and Di Fiore, P. P. (2004). A novel actin barbed-end-capping activity in EPS-8 regulates apical morphogenesis in intestinal cells of Caenorhabditis elegans. Nat. Cell Biol. 6, 1173–1179. - PubMed
- Disanza, A., Carlier, M. F., Stradal, T. E., Didry, D., Frittoli, E., Confalonieri, S., Croce, A., Wehland, J., Di Fiore, P. P., and Scita, G. (2004). Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat. Cell Biol. 6, 1180–1188. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases