Joint modeling of linkage and association: identifying SNPs responsible for a linkage signal - PubMed (original) (raw)
Comparative Study
doi: 10.1086/430277. Epub 2005 Apr 5.
Affiliations
- PMID: 15877278
- PMCID: PMC1196453
- DOI: 10.1086/430277
Comparative Study
Joint modeling of linkage and association: identifying SNPs responsible for a linkage signal
Mingyao Li et al. Am J Hum Genet. 2005 Jun.
Abstract
Once genetic linkage has been identified for a complex disease, the next step is often association analysis, in which single-nucleotide polymorphisms (SNPs) within the linkage region are genotyped and tested for association with the disease. If a SNP shows evidence of association, it is useful to know whether the linkage result can be explained, in part or in full, by the candidate SNP. We propose a novel approach that quantifies the degree of linkage disequilibrium (LD) between the candidate SNP and the putative disease locus through joint modeling of linkage and association. We describe a simple likelihood of the marker data conditional on the trait data for a sample of affected sib pairs, with disease penetrances and disease-SNP haplotype frequencies as parameters. We estimate model parameters by maximum likelihood and propose two likelihood-ratio tests to characterize the relationship of the candidate SNP and the disease locus. The first test assesses whether the candidate SNP and the disease locus are in linkage equilibrium so that the SNP plays no causal role in the linkage signal. The second test assesses whether the candidate SNP and the disease locus are in complete LD so that the SNP or a marker in complete LD with it may account fully for the linkage signal. Our method also yields a genetic model that includes parameter estimates for disease-SNP haplotype frequencies and the degree of disease-SNP LD. Our method provides a new tool for detecting linkage and association and can be extended to study designs that include unaffected family members.
Figures
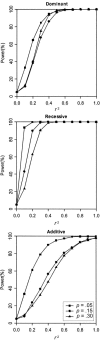
Figure 1
Power to reject linkage equilibrium (
_r_2=0
). Results are based on 2,000 replicates of 500 ASPs. All models have population disease prevalence K = 2% and sibling recurrence-risk ratio
λ_s_=1.1
. Power was assessed at the 5% level.
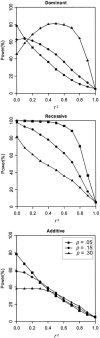
Figure 2
Power to reject complete LD (
_r_2=1
). Results are based on 2,000 replicates of 500 ASPs. All models have population disease prevalence K = 2% and sibling recurrence-risk ratio
λ_s_=1.3
. Power was assessed at the 5% level.
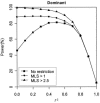
Figure 3
Impact of linkage evidence on test of complete LD. Results are based on 2,000 replicates of 500 ASPs under a dominant model with population disease prevalence K = 2%, allele frequency
p _D_=p _A_=0.30
, and sibling recurrence-risk ratio
λ_s_=1.3
. Power was assessed at the 5% level.
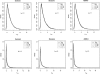
Figure 4
Impact of the number of flanking markers. Results are based on 2,000 replicates of 500 ASPs simulated under an additive model with population disease prevalence K = 2%, allele frequency
p _D_=p _A_=0.15
, and sibling recurrence-risk ratios
λ_s_=1.1
(A) and 1.3 (B). Data were simulated using 10 flanking markers, each with two equally frequent alleles. Intermarker recombination fraction is 0.1. Power was assessed at the 5% level.
Figure 5
Impact of heterozygosity of flanking markers. Results are based on 2,000 replicates of 500 ASPs simulated under an additive model with population disease prevalence K = 2%, allele frequency
p _D_=p _A_=0.15
, and sibling recurrence-risk ratios
λ_s_=1.1
(A) and 1.3 (B). Data were simulated using two flanking markers, each with two, four, or eight equally frequent alleles. Intermarker recombination fraction is 0.1. Power was assessed at the 5% level.
Figure 6
Impact of intermarker recombination of flanking markers. Results are based on 2,000 replicates of 500 ASPs simulated under an additive model with population disease prevalence K = 2%, allele frequency
p _D_=p _A_=0.15
, and sibling recurrence-risk ratios
λ_s_=1.1
(A) and 1.3 (B). Data were simulated using 10 flanking markers, each with four equally frequent alleles. Power was assessed at the 5% level.
Figure 7
Comparison of empirical null distributions. Results are based on 2,000 replicates of 500 ASPs. All models have population disease prevalence K = 2% and allele frequency
p _D_=p _A_=0.15
. The solid line in each plot is the density of the empirical null distribution simulated using true parameter values of the disease model. Dashed lines are density plots of the empirical null distributions generated using the resampling procedures described in the “Methods” section. The empirical null distribution was generated for each level of disease-SNP LD.
Similar articles
- Efficient study designs for test of genetic association using sibship data and unrelated cases and controls.
Li M, Boehnke M, Abecasis GR. Li M, et al. Am J Hum Genet. 2006 May;78(5):778-792. doi: 10.1086/503711. Epub 2006 Mar 20. Am J Hum Genet. 2006. PMID: 16642434 Free PMC article. - Joint Linkage and Association Analysis Using GENEHUNTER-MODSCORE with an Application to Familial Pancreatic Cancer.
Brugger M, Lutz M, Müller-Nurasyid M, Lichtner P, Slater EP, Matthäi E, Bartsch DK, Strauch K. Brugger M, et al. Hum Hered. 2024;89(1):8-31. doi: 10.1159/000535840. Epub 2024 Jan 10. Hum Hered. 2024. PMID: 38198765 - On selecting markers for association studies: patterns of linkage disequilibrium between two and three diallelic loci.
Garner C, Slatkin M. Garner C, et al. Genet Epidemiol. 2003 Jan;24(1):57-67. doi: 10.1002/gepi.10217. Genet Epidemiol. 2003. PMID: 12508256 Review. - Linkage disequilibrium maps and disease-association mapping.
Maniatis N. Maniatis N. Methods Mol Biol. 2007;376:109-21. doi: 10.1007/978-1-59745-389-9_8. Methods Mol Biol. 2007. PMID: 17984541 Review.
Cited by
- Genetics of Smoking Behaviors in American Indians.
Henderson JA, Buchwald DS, Howard BV, Henderson PN, Li Y, Tyndale RF, Amos CI, Gorlova OY; Collaborative to Improve Native Cancer Outcomes (CINCO), a P50 Center for Population Health and Health Disparities program project sponsored by the National Cancer Institute. Henderson JA, et al. Cancer Epidemiol Biomarkers Prev. 2020 Nov;29(11):2180-2186. doi: 10.1158/1055-9965.EPI-20-0026. Epub 2020 Aug 27. Cancer Epidemiol Biomarkers Prev. 2020. PMID: 32855268 Free PMC article. - Evidence for association of bipolar disorder to haplotypes in the 22q12.3 region near the genes stargazin, IFT27 and parvalbumin.
Nissen S, Liang S, Shehktman T, Kelsoe JR; Bipolar Genome Study (BiGS); Greenwood TA, Nievergelt CM, McKinney R, Shilling PD, Smith EN, Schork NJ, Bloss CS, Nurnberger JI Jr, Edenberg HJ, Foroud T, Koller DL, Gershon ES, Liu C, Badner JA, Scheftner WA, Lawson WB, Nwulia EA, Hipolito M, Coryell W, Rice J, Byerley W, McMahon FJ, Berrettini WH, Potash JB, Zandi PP, Mahon PB, McInnis MG, Zöllner S, Zhang P, Craig DW, Szelinger S, Barrett TB, Schulze TG. Nissen S, et al. Am J Med Genet B Neuropsychiatr Genet. 2012 Dec;159B(8):941-50. doi: 10.1002/ajmg.b.32099. Epub 2012 Oct 4. Am J Med Genet B Neuropsychiatr Genet. 2012. PMID: 23038240 Free PMC article. - HTR1A a novel type 1 diabetes susceptibility gene on chromosome 5p13-q13.
Asad S, Nikamo P, Gyllenberg A, Bennet H, Hansson O, Wierup N; Diabetes Incidence in Sweden Study Group; Carlsson A, Forsander G, Ivarsson SA, Larsson H, Lernmark Å, Lindblad B, Ludvigsson J, Marcus C, Rønningen KS, Nerup J, Pociot F, Luthman H, Fex M, Kockum I. Asad S, et al. PLoS One. 2012;7(5):e35439. doi: 10.1371/journal.pone.0035439. Epub 2012 May 1. PLoS One. 2012. PMID: 22563461 Free PMC article. - Strong association of the Y402H variant in complement factor H at 1q32 with susceptibility to age-related macular degeneration.
Zareparsi S, Branham KE, Li M, Shah S, Klein RJ, Ott J, Hoh J, Abecasis GR, Swaroop A. Zareparsi S, et al. Am J Hum Genet. 2005 Jul;77(1):149-53. doi: 10.1086/431426. Epub 2005 May 13. Am J Hum Genet. 2005. PMID: 15895326 Free PMC article. - On the validity of the likelihood ratio test and consistency of resulting parameter estimates in joint linkage and linkage disequilibrium analysis under improperly specified parametric models.
Hiekkalinna T, Göring HH, Terwilliger JD. Hiekkalinna T, et al. Ann Hum Genet. 2012 Jan;76(1):63-73. doi: 10.1111/j.1469-1809.2011.00683.x. Epub 2011 Nov 14. Ann Hum Genet. 2012. PMID: 22082140 Free PMC article.
References
Electronic-Database Information
- University of Michigan Center for Statistical Genetics, http://csg.sph.umich.edu/
References
- Baum LE (1972) An inequality and associated maximization technique in statistical estimation for probabilistic functions of Markov processes. Inequalities 3:1–8
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials