Pericentromeric regions of soybean (Glycine max L. Merr.) chromosomes consist of retroelements and tandemly repeated DNA and are structurally and evolutionarily labile - PubMed (original) (raw)
Comparative Study
Pericentromeric regions of soybean (Glycine max L. Merr.) chromosomes consist of retroelements and tandemly repeated DNA and are structurally and evolutionarily labile
Jer-Young Lin et al. Genetics. 2005 Jul.
Abstract
Little is known about the physical makeup of heterochromatin in the soybean (Glycine max L. Merr.) genome. Using DNA sequencing and molecular cytogenetics, an initial analysis of the repetitive fraction of the soybean genome is presented. BAC 076J21, derived from linkage group L, has sequences conserved in the pericentromeric heterochromatin of all 20 chromosomes. FISH analysis of this BAC and three subclones on pachytene chromosomes revealed relatively strict partitioning of the heterochromatic and euchromatic regions. Sequence analysis showed that this BAC consists primarily of repetitive sequences such as a 102-bp tandem repeat with sequence identity to a previously characterized approximately 120-bp repeat (STR120). Fragments of Calypso-like retroelements, a recently inserted SIRE1 element, and a SIRE1 solo LTR were present within this BAC. Some of these sequences are methylated and are not conserved outside of G. max and G. soja, a close relative of soybean, except for STR102, which hybridized to a restriction fragment from G. latifolia. These data present a picture of the repetitive fraction of the soybean genome that is highly concentrated in the pericentromeric regions, consisting of rapidly evolving tandem repeats with interspersed retroelements.
Figures
Figure 1.—
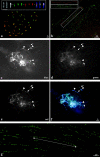
Fluorescence in situ hybridization analysis of BACs and subclones to chromosomes and extended DNA fibers of soybean. (a) FISH of BAC 076J21 (green) to mitotic chromosomes (red). (Inset) Three chromosomes from another preparation showing, from left to right: DAPI stained (black and white), 076J21 (green), 09M21 (red), and merged. (b) Fiber-FISH of BACs 076J21 (red) and 09M21 (green) to DNA fibers of soybean showing little overlap in FISH signal. (Inset) An image of a single fiber with the two color channels shown separately. (c–f) FISH to pachytene chromosomes of soybean with subclones of BAC 076J21. Arrows indicate heterochromatic regions and arrowheads indicate centromeric heterochromatin. (c) DAPI-stained chromosomes. (d) STR102 [2_P01]. (e) SIRE 1 [1_L22] and Calypso 5-1 [1_E15] pooled. (f) Merged image. (g) Fiber-FISH analysis of STR102 [2_P01] on extended DNA fibers of soybean showing long interrupted arrays of STR102 [2_P01]. Line with arrows indicates a 435.6-kb cluster of repeats.
Figure 2.—
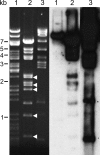
Southern analysis of shared sequences between BACs 09M21 and 076J21. (Left) Ethidium-bromide-stained gel of restriction-digested BACs before blotting. Lane 1, BAC 123E07; lane 2, BAC 09M21; lane 3, BAC 076J21. (Right) Hybridization of 076J21 to gel blot. BAC 123E07 is derived from a euchromatic region of soybean and only the BAC vector cross-hybridizes (∼7.4 kb). BAC 09M21 shares some sequences with 076J21 but some restriction bands (arrowheads) show little cross-hybridization.
Figure 3.—
Sequence analysis of BAC 076J21. (a) Schematic of BAC 076J21 (79,623 bp). Black bars above sequence diagram are the subclones used for Southern analysis and FISH. Across the top, a 200-bp sliding window (_x_-axis) was used to map the number of BLASTN hits (_y_-axis) from a search against the soybean GSS sequences along the length of 076J21. FGENESH-predicted genes are shown with arrows along the bottom of the diagram. (b) Digital mapping of 2_P01 [STR102] onto BAC 076J21 using FISH. Three clusters are seen, two of which (arrows) may border the SIRE1 element. The other (star) did not assemble into the sequence scaffold. (c) ClustalW was used to align two representatives of the STR102 repeat (STR102_1 and STR102_4) with five STR120 members from GenBank. Asterisks denote complete identity among all seven sequences at a nucleotide position. (d) A dot plot was made using Dotter (S
onnhammer
and D
urbin
- with a word size of 20 nt of 076J21 against itself. A schematic of the BAC with internal structures is shown on both axes. The LTRs of the SIRE1 element are indicated with green arrows and the solo LTR with a red arrow.
Figure 4.—
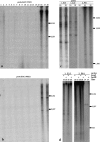
Southern analysis of conservation of BACs 076J21 and 09M21 and conservation and methylation of subclones of 076J21. Lane numbers for all a–d refer to the species in Table 1. (a) BAC 076J21 and (b) 09M21 probed against the Glycine species blot. (c) Subclones of 076J21 probed against the Glycine species blots (only lanes showing hybridization are shown). (d) Methylation of 076J21 subclones containing Calypso 5-1 (1E_15) and STR102 (2P_01) were tested by probing against genomic DNA digested with either _Msp_I or _Hpa_II.
Similar articles
- Major repeat components covering one-third of the ginseng (Panax ginseng C.A. Meyer) genome and evidence for allotetraploidy.
Choi HI, Waminal NE, Park HM, Kim NH, Choi BS, Park M, Choi D, Lim YP, Kwon SJ, Park BS, Kim HH, Yang TJ. Choi HI, et al. Plant J. 2014 Mar;77(6):906-16. doi: 10.1111/tpj.12441. Epub 2014 Feb 24. Plant J. 2014. PMID: 24456463 - Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison.
Du J, Tian Z, Hans CS, Laten HM, Cannon SB, Jackson SA, Shoemaker RC, Ma J. Du J, et al. Plant J. 2010 Aug;63(4):584-98. doi: 10.1111/j.1365-313X.2010.04263.x. Plant J. 2010. PMID: 20525006 - Segmental duplications within the Glycine max genome revealed by fluorescence in situ hybridization of bacterial artificial chromosomes.
Pagel J, Walling JG, Young ND, Shoemaker RC, Jackson SA. Pagel J, et al. Genome. 2004 Aug;47(4):764-8. doi: 10.1139/g04-025. Genome. 2004. PMID: 15284882 - Tandemly repeated DNA sequences and centromeric chromosomal regions of Arabidopsis species.
Heslop-Harrison JS, Brandes A, Schwarzacher T. Heslop-Harrison JS, et al. Chromosome Res. 2003;11(3):241-53. doi: 10.1023/a:1022998709969. Chromosome Res. 2003. PMID: 12769291 Review. - Complex structure of knobs and centromeric regions in maize chromosomes.
Ananiev EV, Phillips RL, Rines HW. Ananiev EV, et al. Tsitol Genet. 2000 Mar-Apr;34(2):11-5. Tsitol Genet. 2000. PMID: 10857197 Review.
Cited by
- Fluorescence in situ hybridization-based karyotyping of soybean translocation lines.
Findley SD, Pappas AL, Cui Y, Birchler JA, Palmer RG, Stacey G. Findley SD, et al. G3 (Bethesda). 2011 Jul;1(2):117-29. doi: 10.1534/g3.111.000034. Epub 2011 Jul 1. G3 (Bethesda). 2011. PMID: 22384324 Free PMC article. - A fluorescence in situ hybridization system for karyotyping soybean.
Findley SD, Cannon S, Varala K, Du J, Ma J, Hudson ME, Birchler JA, Stacey G. Findley SD, et al. Genetics. 2010 Jul;185(3):727-44. doi: 10.1534/genetics.109.113753. Epub 2010 Apr 26. Genetics. 2010. PMID: 20421607 Free PMC article. - Functional centromeres in soybean include two distinct tandem repeats and a retrotransposon.
Tek AL, Kashihara K, Murata M, Nagaki K. Tek AL, et al. Chromosome Res. 2010 Apr;18(3):337-47. doi: 10.1007/s10577-010-9119-x. Epub 2010 Mar 5. Chromosome Res. 2010. PMID: 20204495 - Sequence conservation of homeologous bacterial artificial chromosomes and transcription of homeologous genes in soybean (Glycine max L. Merr.).
Schlueter JA, Scheffler BE, Schlueter SD, Shoemaker RC. Schlueter JA, et al. Genetics. 2006 Oct;174(2):1017-28. doi: 10.1534/genetics.105.055020. Epub 2006 Aug 3. Genetics. 2006. PMID: 16888343 Free PMC article. - Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing.
Schlueter JA, Lin JY, Schlueter SD, Vasylenko-Sanders IF, Deshpande S, Yi J, O'Bleness M, Roe BA, Nelson RT, Scheffler BE, Jackson SA, Shoemaker RC. Schlueter JA, et al. BMC Genomics. 2007 Sep 19;8:330. doi: 10.1186/1471-2164-8-330. BMC Genomics. 2007. PMID: 17880721 Free PMC article.
References
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. - PubMed
- Aragon-Alcaide, L., T. Miller, T. Schwarzacher, S. Reader and G. Moore, 1996. A cereal centromeric sequence. Chromosoma 105: 261–268. - PubMed
- Arumuganathan, K., and E. D. Earle, 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 229–241.
- Bennetzen, J. L., 1996. The contributions of retroelements to plant genome organization, function and evolution. Trends Microbiol. 4: 347–353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources