Cooperative interaction of human XPA stabilizes and enhances specific binding of XPA to DNA damage - PubMed (original) (raw)
Cooperative interaction of human XPA stabilizes and enhances specific binding of XPA to DNA damage
Yu Liu et al. Biochemistry. 2005.
Abstract
Human xeroderma pigmentosum group A (XPA) is an essential protein for nucleotide excision repair (NER). We have previously reported that XPA forms a homodimer in the absence of DNA. However, what oligomeric forms of XPA are involved in DNA damage recognition and how the interaction occurs in terms of biochemical understanding remain unclear. Using the homogeneous XPA protein purified from baculovirus-infected Sf21 insect cells and the methods of gel mobility shift assays, gel filtration chromatography, and UV-cross-linking, we demonstrated that both monomeric and dimeric XPA bound to the DNA adduct of N-acetyl-2-aminofluorene (AAF), while showing little affinity for nondamaged DNA. The binding occurred in a sequential and protein concentration-dependent manner. At relatively low-protein concentrations, XPA formed a complex with DNA adduct as a monomer, while at the higher concentrations, an XPA dimer was involved in the specific binding. Results from fluorescence spectroscopic and competitive binding analyses indicated that the specific binding of XPA to the adduct was significantly facilitated and stabilized by the presence of the second XPA in a positive cooperative manner. This cooperative binding exhibited a Hill coefficient of 1.9 and the step binding constants of K(1) = 1.4 x 10(6) M(-)(1) and K(2) = 1.8 x 10(7) M(-)(1). When interaction of XPA and RPA with DNA was studied, even though binding of RPA-XPA complex to adducted DNA was observed, the presence of RPA had little effect on the overall binding efficiency. Our results suggest that the dominant form for XPA to efficiently bind to DNA damage is the XPA dimer. We hypothesized that the concentration-dependent formation of different types of XPA-damaged DNA complex may play a role in cellular regulation of XPA activity.
Figures
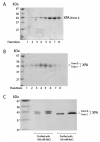
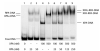
FIGURE 1:
SDS-PAGE of purified XPA. [His]6-XPA expressed from the sf21 cells was purified either with 100 mM (A) or 500 mM (B) (see Materials and Methods) and analyzed on an 8% SDS-PAGE stained with Coomassie Blue. (C) The comparison of the XPAs purified from different experimental conditions.
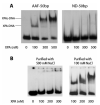
FIGURE 2:
Interactions of XPA with AAF-DNA substrate. (A) XPA binds to a 50-bp AAF adduct showing two well-resolved XPA-DNA bands. The XPA of varying concentrations were incubated with AAF-50-bp or nondamaged (ND) substrate (2 nM) at 30 °C for 20 min in 20 μL binding buffer (20 mM Hepes, pH 7.9, 75 mM KCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 5% glycerol, and 100 mg/mL BSA). The mixtures were loaded onto a 3.5% native polyacrylamide gel and electrophoresed at 4 °C. (B) The same gel mobility shift assays for the binding of XPA purified with 100 mM and 500 mM NaCl to AAF-50-bp substrate, respectively.
FIGURE 3:
Gel filtration analysis of XPA binding to AAF-50-bp. (A) XPA protein at various concentrations (100-1500 nM) was incubated with 4 nM 5́ terminally [32P]-labeled AAF-50-bp DNA substrate at 30 °C for 20 min in DNA binding buffer (20 mM Hepes, pH 7.9, 75 mM KCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 5% glycerol, and 100 mg/mL BSA). The mixture was then loaded onto a Superdex-200 HR column running with binding buffer for separation. Fractions (200 μL) were collected and counted for radioactivity. The radioactive intensities of fractions were plotted against the corresponding retention volume. (B) The molecular weights of the formed XPA-DNA complexes were determined based on a set of molecular mass markers (ribonuclease A, 13.7 kDa; chymotrysinogen A, 25 kDa; ovalbumin, 43 kDa; albumin, 67 kDa; aldolase, 158 kDa; catalase, 232 kDa; ferritin, 440 kDa; and thyroglobulin, 669 kDa) and gel-filtrated under the same condition as the XPA samples. Data for these markers were fitted with linear regression method. At 100 nM of XPA, the retention volume corresponds to a molecular weight of 65 kDa, while at the concentrations g500 nM, the retention volume of XPA-DNA complex indicates a molecular weight of 98 kDa.
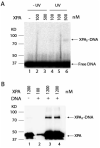
FIGURE 4:
Cross-linking of XPA-DNA complex by UV irradiation. (A) 32P terminally labeled AAF-DNA substrate was incubated with the indicated concentrations of XPA and then subjected to UV irradiation (254 nm) at 14 J m-2 s-1. The samples were analyzed on 8% SDS-PAGE. The radioactivity of the gel was detected by phosphorimage scanner. (B) The samples were prepared as the same as in panel A except that the UV irradiation was performed at 312 nm. After 8% SDS-PAGE separation, the gel was subjected to Western blotting with anti-XPA antibody.
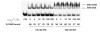
FIGURE 5:
Competitive binding of nondamaged and damaged DNA to XPA. The XPA protein (100 nM and 500 nM) was incubated with [32P]-labeled AAF-50-bp DNA substrate at 30 °C for 20 min in the binding buffer in the presence of nondamaged 50-bp DNA (nonradioactive) at various concentrations or ratios of nondamaged DNA to damaged DNA. Samples were then analyzed by gel mobility shift assays at 4 °C. The XPA-DNA complex formation was quantified as the percentage of DNA bound to XPA.
FIGURE 6:
Fluorescence anisotropy measurements of XPA binding to 5́F-AAF-50-bp. (A) Binding isotherm was generated by titrating the 5́ terminally labeled AAF-50-bp DNA (10 nM) with XPA protein in 200 μL of binding buffer. The anisotropy was recorded at an emission wavelength of 520 nm with an excitation wavelength of 492 nm. The anisotropy data was normalized by subtracting the initial value of free DNA. The error bars indicate the standard deviation of at least three independent experiments. The solid line represents the best fitting of the binding isotherm with a two-binding model by nonlinear least-squares strategy as described in Materials and Methods. From the fitting, the binding constants were determined as _K_1) 1.4 × 106 M-1 and _K_2) 1.8 × 107 M-1. (B) The fluorescence anisotropy data were analyzed by Hill plot, which gave a Hill coefficient of 1.91, an indication of possible cooperative interaction of XPA with DNA adduct.
FIGURE 7:
Interaction of RPA and XPA with AAF-DNA substrate. The XPA at 100 and 500 nM were incubated with AAF-50-bp DNA in the presence (lanes 6-9) and absence (lane 4-5) of RPA of various concentrations at 30 °C for 20 min in binding buffer. Samples were analyzed by gel mobility shift assays on a 3.5% native polyacrylamide gel at 4 °C. Lanes 2-3, RPA binds to the DNA substrate in the absence of XPA.
Similar articles
- Binding of XPA and RPA to damaged DNA investigated by fluorescence anisotropy.
Hey T, Lipps G, Krauss G. Hey T, et al. Biochemistry. 2001 Mar 6;40(9):2901-10. doi: 10.1021/bi002166i. Biochemistry. 2001. PMID: 11258902 - Specific and efficient binding of xeroderma pigmentosum complementation group A to double-strand/single-strand DNA junctions with 3'- and/or 5'-ssDNA branches.
Yang Z, Roginskaya M, Colis LC, Basu AK, Shell SM, Liu Y, Musich PR, Harris CM, Harris TM, Zou Y. Yang Z, et al. Biochemistry. 2006 Dec 26;45(51):15921-30. doi: 10.1021/bi061626q. Epub 2006 Dec 19. Biochemistry. 2006. PMID: 17176115 Free PMC article. - Dimerization of human XPA and formation of XPA2-RPA protein complex.
Yang ZG, Liu Y, Mao LY, Zhang JT, Zou Y. Yang ZG, et al. Biochemistry. 2002 Oct 29;41(43):13012-20. doi: 10.1021/bi026064z. Biochemistry. 2002. PMID: 12390028 Free PMC article. - Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair.
Thoma BS, Vasquez KM. Thoma BS, et al. Mol Carcinog. 2003 Sep;38(1):1-13. doi: 10.1002/mc.10143. Mol Carcinog. 2003. PMID: 12949838 Review. - Interactions of mammalian proteins with cisplatin-damaged DNA.
Turchi JJ, Henkels KM, Hermanson IL, Patrick SM. Turchi JJ, et al. J Inorg Biochem. 1999 Oct;77(1-2):83-7. doi: 10.1016/s0162-0134(99)00145-2. J Inorg Biochem. 1999. PMID: 10626358 Review.
Cited by
- Single molecule analysis reveals monomeric XPA bends DNA and undergoes episodic linear diffusion during damage search.
Beckwitt EC, Jang S, Carnaval Detweiler I, Kuper J, Sauer F, Simon N, Bretzler J, Watkins SC, Carell T, Kisker C, Van Houten B. Beckwitt EC, et al. Nat Commun. 2020 Mar 13;11(1):1356. doi: 10.1038/s41467-020-15168-1. Nat Commun. 2020. PMID: 32170071 Free PMC article. - XPA expression is a predictive marker of the effectiveness of neoadjuvant chemotherapy for locally advanced uterine cervical cancer.
Wada T, Fukuda T, Shimomura M, Inoue Y, Kawanishi M, Tasaka R, Yasui T, Ikeda K, Sumi T. Wada T, et al. Oncol Lett. 2018 Mar;15(3):3766-3771. doi: 10.3892/ol.2018.7810. Epub 2018 Jan 16. Oncol Lett. 2018. PMID: 29556276 Free PMC article. - RPA and XPA interaction with DNA structures mimicking intermediates of the late stages in nucleotide excision repair.
Krasikova YS, Rechkunova NI, Maltseva EA, Lavrik OI. Krasikova YS, et al. PLoS One. 2018 Jan 10;13(1):e0190782. doi: 10.1371/journal.pone.0190782. eCollection 2018. PLoS One. 2018. PMID: 29320546 Free PMC article. - Xeroderma Pigmentosa Group A (XPA), Nucleotide Excision Repair and Regulation by ATR in Response to Ultraviolet Irradiation.
Musich PR, Li Z, Zou Y. Musich PR, et al. Adv Exp Med Biol. 2017;996:41-54. doi: 10.1007/978-3-319-56017-5_4. Adv Exp Med Biol. 2017. PMID: 29124689 Free PMC article. Review. - Analysis of DNA binding by human factor xeroderma pigmentosum complementation group A (XPA) provides insight into its interactions with nucleotide excision repair substrates.
Sugitani N, Voehler MW, Roh MS, Topolska-Woś AM, Chazin WJ. Sugitani N, et al. J Biol Chem. 2017 Oct 13;292(41):16847-16857. doi: 10.1074/jbc.M117.800078. Epub 2017 Aug 31. J Biol Chem. 2017. PMID: 28860187 Free PMC article.
References
- Petit C, Sancar A. Nucleotide excision repair: from E. coli to man. Biochimie. 1999;81:15–25. - PubMed
- Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. - PubMed
- Thoma BS, Vasquez KM. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol. Carcinog. 2003:1–13. - PubMed
- Hanawalt PC. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. - PubMed
- de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 ES009127/ES/NIEHS NIH HHS/United States
- ES00318/ES/NIEHS NIH HHS/United States
- R01 CA086927/CA/NCI NIH HHS/United States
- R56 ES009127/ES/NIEHS NIH HHS/United States
- R56 CA086927/CA/NCI NIH HHS/United States
- CA86927/CA/NCI NIH HHS/United States
- ES09127/ES/NIEHS NIH HHS/United States
- K02 ES000318/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources