Autophagy promotes MHC class II presentation of peptides from intracellular source proteins - PubMed (original) (raw)
. 2005 May 31;102(22):7922-7.
doi: 10.1073/pnas.0501190102. Epub 2005 May 13.
Oliver Schoor, Rainer Fischer, Michael Reich, Marianne Kraus, Margret Müller, Katharina Kreymborg, Florian Altenberend, Jens Brandenburg, Hubert Kalbacher, Roland Brock, Christoph Driessen, Hans-Georg Rammensee, Stefan Stevanovic
Affiliations
- PMID: 15894616
- PMCID: PMC1142372
- DOI: 10.1073/pnas.0501190102
Autophagy promotes MHC class II presentation of peptides from intracellular source proteins
Jörn Dengjel et al. Proc Natl Acad Sci U S A. 2005.
Abstract
MHC-peptide complexes mediate key functions in adaptive immunity. In a classical view, MHC-I molecules present peptides from intracellular source proteins, whereas MHC-II molecules present antigenic peptides from exogenous and membrane proteins. Nevertheless, substantial crosstalk between these two pathways has been observed. We investigated the influence of autophagy on the MHC-II ligandome and demonstrated that peptide presentation is altered considerably upon induction of autophagy. The presentation of peptides from intracellular and lysosomal source proteins was strongly increased on MHC-II in contrast with peptides from membrane and secreted proteins. In addition, autophagy influenced the MHC-II antigen-processing machinery. Our study illustrates a profound influence of autophagy on the class II peptide repertoire and suggests that this finding has implications for the regulation of CD4(+) T cell-mediated processes.
Figures
Fig. 1.
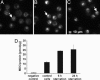
Starvation enhances the level of autophagic vacuoles. Autophagic vacuoles were stained with the specific dye MDC (13) and analyzed by fluorescence microscopy or fluorescence spectroscopy. Awells cells were incubated for 24 h in DMEM (control cells) (A), 6 h HBSS (B), or 24 h HBSS (starved cells) (C), subsequently, for 10 min with MDC, washed, and immediately analyzed by fluorescence microscopy. Autophagic vacuoles are marked with an arrow. (D) Intracellular MDC measurement by fluorescence spectroscopy; unstained cells were used as negative control.
Fig. 2.
Altered peptide presentation on HLA-DR under starvation. Displayed are the relative intensity ratios of HLA-DR-eluted peptides from starved cells (6 and 24 h) and control cells as assessed by LC-MS. Peptides were quantified by their relative peak heights in mass spectra and grouped according to the cellular localization of their source proteins: membrane plus secreted proteins and intracellular plus lysosomal proteins. Data of serial LC-MS runs were normalized to the abundant peptide LSSWTAADTAAQITQR, which showed only marginal differences in presentation levels (Table 1). Horizontal bars indicate the mean intensity ratios for each group. Marked in a box are the four peptides that showed the highest presentation levels after 24 h of starvation. Their source proteins are localized in the nucleus and in lysosomes.
Fig. 3.
Changes of lysosomal protease composition during autophagy. (A) Affinity labeling of active cathepsins. Endocytic extracts were generated from control cells, cells after 6 h and 24 h of starvation, and human peripheral blood monoyctes, respectively, by differential centrifugation as reported in refs. and . Five-microgram total endocytic protein (1.5 μg in monocytes) were either directly incubated with the active site-restricted biotinylated affinity label DCG-0N as described (lanes: 2, control cells; 3, 6 h of starvation; 4, 24 h of starvation; and 9, monocytes) or were subjected to 95°C as negative control (lane 1). In addition, control cells were incubated with the CatS-inhibitor LHVS (25 nM), the CatB-inhibitor Ca074 (1 μM), the pan-cysteine protease inhibitors leupeptin (1 mM), or E64 (25 μM) (lanes 5–8) for 45 min at 37°C before labeling as further controls. Active cathepsins were visualized after resolution by SDS/PAGE by streptavidin-HRP blot: CatZ, CatB, CatH, and CatS at 36, 33, 30, and 28 kDa, respectively. (B) Cathepsin polypeptides probed by Western blot. Identical amounts of total cellular protein from control cells (lane 1) and cells undergoing autophagy (6 and 24 h of starvation, respectively; lane 2 and lane 3) were probed for CatS, CatC, CatD, CatH, β-actin, and LAMP-1 by Western blot.
Fig. 4.
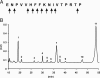
MBP83–99 digestion with lysosomal extracts from control cells and cells undergoing autophagy. (A) Preferential detected cleavage sites. MBP83–99 was incubated at pH 5.4 with lysosomal extracts from control cells and cells undergoing 24 h autophagy for 3 h. Breakdown products were subsequently separated by RP-HLPC and analyzed by MALDI-MS and Edman microsequencing. (B) RP-HLPC chromatogram of control cell MBP breakdown products at 214 nm. The annotated peaks correspond to the major breakdown products of MBP83–99 as identified by MALDI MS and Edman microsequencing. The corresponding chromatogram of autophagic cells is not shown.
Comment in
- Autophagy and intracellular surveillance: Modulating MHC class II antigen presentation with stress.
Crotzer VL, Blum JS. Crotzer VL, et al. Proc Natl Acad Sci U S A. 2005 May 31;102(22):7779-80. doi: 10.1073/pnas.0503088102. Epub 2005 May 23. Proc Natl Acad Sci U S A. 2005. PMID: 15911750 Free PMC article. No abstract available.
Similar articles
- Autophagy Beyond Intracellular MHC Class II Antigen Presentation.
Münz C. Münz C. Trends Immunol. 2016 Nov;37(11):755-763. doi: 10.1016/j.it.2016.08.017. Epub 2016 Sep 22. Trends Immunol. 2016. PMID: 27667710 Review. - Autophagy and intracellular surveillance: Modulating MHC class II antigen presentation with stress.
Crotzer VL, Blum JS. Crotzer VL, et al. Proc Natl Acad Sci U S A. 2005 May 31;102(22):7779-80. doi: 10.1073/pnas.0503088102. Epub 2005 May 23. Proc Natl Acad Sci U S A. 2005. PMID: 15911750 Free PMC article. No abstract available. - The formation of immunogenic major histocompatibility complex class II-peptide ligands in lysosomal compartments of dendritic cells is regulated by inflammatory stimuli.
Inaba K, Turley S, Iyoda T, Yamaide F, Shimoyama S, Reis e Sousa C, Germain RN, Mellman I, Steinman RM. Inaba K, et al. J Exp Med. 2000 Mar 20;191(6):927-36. doi: 10.1084/jem.191.6.927. J Exp Med. 2000. PMID: 10727455 Free PMC article. - Autophagy and MHC-restricted antigen presentation.
Valečka J, Almeida CR, Su B, Pierre P, Gatti E. Valečka J, et al. Mol Immunol. 2018 Jul;99:163-170. doi: 10.1016/j.molimm.2018.05.009. Epub 2018 May 19. Mol Immunol. 2018. PMID: 29787980 Review. - Autophagy in MHC class II antigen processing.
Strawbridge AB, Blum JS. Strawbridge AB, et al. Curr Opin Immunol. 2007 Feb;19(1):87-92. doi: 10.1016/j.coi.2006.11.009. Epub 2006 Nov 28. Curr Opin Immunol. 2007. PMID: 17129719 Review.
Cited by
- Chikungunya virus-induced autophagy delays caspase-dependent cell death.
Joubert PE, Werneke SW, de la Calle C, Guivel-Benhassine F, Giodini A, Peduto L, Levine B, Schwartz O, Lenschow DJ, Albert ML. Joubert PE, et al. J Exp Med. 2012 May 7;209(5):1029-47. doi: 10.1084/jem.20110996. Epub 2012 Apr 16. J Exp Med. 2012. PMID: 22508836 Free PMC article. - Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells.
Ireland JM, Unanue ER. Ireland JM, et al. J Exp Med. 2011 Dec 19;208(13):2625-32. doi: 10.1084/jem.20110640. Epub 2011 Dec 12. J Exp Med. 2011. PMID: 22162830 Free PMC article. - Macroautophagy substrates are loaded onto MHC class II of medullary thymic epithelial cells for central tolerance.
Aichinger M, Wu C, Nedjic J, Klein L. Aichinger M, et al. J Exp Med. 2013 Feb 11;210(2):287-300. doi: 10.1084/jem.20122149. Epub 2013 Feb 4. J Exp Med. 2013. PMID: 23382543 Free PMC article. - Hypothermia-Mediated Apoptosis and Inflammation Contribute to Antioxidant and Immune Adaption in Freshwater Drum, Aplodinotus grunniens.
Chen J, Li H, Xu P, Tang Y, Su S, Liu G, Wu N, Xue M, Yu F, Feng W, Song C, Wen H. Chen J, et al. Antioxidants (Basel). 2022 Aug 26;11(9):1657. doi: 10.3390/antiox11091657. Antioxidants (Basel). 2022. PMID: 36139731 Free PMC article. - Involvement of autophagy in viral infections: antiviral function and subversion by viruses.
Espert L, Codogno P, Biard-Piechaczyk M. Espert L, et al. J Mol Med (Berl). 2007 Aug;85(8):811-23. doi: 10.1007/s00109-007-0173-6. Epub 2007 Mar 6. J Mol Med (Berl). 2007. PMID: 17340132 Free PMC article. Review.
References
- Rammensee, H. G., Falk, K. & Rotzschke, O. (1993) Curr. Opin. Immunol. 5, 35–44. - PubMed
- Villadangos, J. A. (2001) Mol. Immunol. 38, 329–346. - PubMed
- Rammensee, H. G., Bachmann, J. & Stevanovic, S. (1997) MHC Ligands and Peptide Motifs (Springer-Verlag, Heidelberg, Germany).
- Princiotta, M. F., Finzi, D., Qian, S. B., Gibbs, J., Schuchmann, S., Buttgereit, F., Bennink, J. R. & Yewdell, J. W. (2003) Immunity 18, 343–354. - PubMed
- Neefjes, J. (1999) Eur. J. Immunol. 29, 1421–1425. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials