Targeting quantum dots to surface proteins in living cells with biotin ligase - PubMed (original) (raw)
Comparative Study
. 2005 May 24;102(21):7583-8.
doi: 10.1073/pnas.0503125102. Epub 2005 May 16.
Affiliations
- PMID: 15897449
- PMCID: PMC1129026
- DOI: 10.1073/pnas.0503125102
Comparative Study
Targeting quantum dots to surface proteins in living cells with biotin ligase
Mark Howarth et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Escherichia coli biotin ligase site-specifically biotinylates a lysine side chain within a 15-amino acid acceptor peptide (AP) sequence. We show that mammalian cell surface proteins tagged with AP can be biotinylated by biotin ligase added to the medium, while endogenous proteins remain unmodified. The biotin group then serves as a handle for targeting streptavidin-conjugated quantum dots (QDs). This labeling method helps to address the two major deficiencies of antibody-based labeling, which is currently the most common method for targeting QDs to cells: the size of the QD conjugate after antibody attachment and the instability of many antibody-antigen interactions. To demonstrate the versatility of our method, we targeted QDs to cell surface cyan fluorescent protein and epidermal growth factor receptor in HeLa cells and to alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPA) receptors in neurons. Labeling requires only 2 min, is extremely specific for the AP-tagged protein, and is highly sensitive. We performed time-lapse imaging of single QDs bound to AMPA receptors in neurons, and we compared the trafficking of different AMPA receptor subunits by using two-color pulse-chase labeling.
Figures
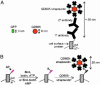
Fig. 1.
General strategy for targeting QDs to cell surface proteins. (A) The size of GFP is compared with unconjugated QD605 and QD605-streptavidin conjugated to biotinylated secondary antibody and primary antibody, as used to label a cell surface protein. (B) The AP, GLNDIFEAQKIEWHE (shown in pink), is genetically encoded at the N terminus or C terminus of the protein of interest. Recombinantly expressed biotin ligase (BirA) is added to the cell medium with ATP and biotin, biotinylating AP. Excess biotin is removed by washing. Streptavidin-conjugated QDs are added to bind the biotinylated surface proteins. B, biotin.
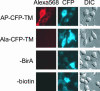
Fig. 2.
Specific labeling of a cell surface protein by BirA shown by imaging. HeLa cells expressing AP-CFP-TM were biotinylated with BirA for 10 min. Biotinylated proteins at the cell surface were detected with streptavidin-Alexa Fluor 568. Controls are shown with a point mutation in AP (Ala-CFP-TM) or with BirA or biotin omitted from the labeling reaction.
Fig. 3.
Specific labeling of cell surface proteins by BirA shown by immunoblot. HeLa cells transfected with AP-CFP-TM were biotinylated with BirA for 5 min. Total cell lysates were blotted with streptavidin-horseradish peroxidase to detect biotinylated proteins. Results are shown for: lane 1, a point mutation in AP (Ala-CFP-TM); lane 2, without BirA; lane 3, without biotin; lane 4, with all components present. Endogenous biotinylated proteins are labeled with arrows and serve as a control for equivalent loading in each lane.
Fig. 4.
QD labeling of cell surface proteins in HeLa cells. HeLa cells expressing AP-CFP-TM were biotinylated with BirA for 5 min. Biotinylation was detected with streptavidin-QD605. A control is shown with a point mutation in AP (Ala-CFP-TM). The QD605 signal is shown with 3-s and 0.2-s exposure.
Fig. 5.
Targeting QDs to AP-tagged AMPA receptors in neurons. Neurons transfected with AP-GluR2 were labeled with BirA for 5 min and then incubated with streptavidin-QD605. A control is shown with a point mutation in AP (Ala-GluR2). CFP was used as a cotransfection marker.
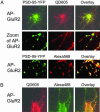
Fig. 6.
The effect of QD size on surface labeling in neurons. (A) Neurons expressing AP-GluR2 and the synaptic marker PSD-95-YFP were labeled with BirA for 10 min at 37°C. After biotinylation, neurons were stained with streptavidin-QD605. (Middle) The staining at higher zoom. (Bottom) The same experiment, except the biotinylation was detected with streptavidin-Alexa Fluor 568 instead of streptavidin-QD605. (B) Neurons expressing AP-GluR2 were labeled with BirA for 10 min at 37°C. They were then incubated simultaneously with streptavidin-Alexa Fluor 488 and streptavidin-QD605.
Fig. 7.
Pulse–chase labeling of AMPA receptor subunits. Neurons were transfected with AP-GluR1 or AP-GluR2 and labeled with BirA for 10 min at 37°C followed by streptavidin-Alexa Fluor 488 for 10 min at room temperature. After 3 min at room temperature with 200 μM glycine, the neurons were again incubated with BirA for 10 min at 37°C and streptavidin-QD605 for 10 min at 4°C before imaging.
Similar articles
- Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase.
Chen I, Howarth M, Lin W, Ting AY. Chen I, et al. Nat Methods. 2005 Feb;2(2):99-104. doi: 10.1038/nmeth735. Epub 2005 Jan 21. Nat Methods. 2005. PMID: 15782206 - Phage display evolution of a peptide substrate for yeast biotin ligase and application to two-color quantum dot labeling of cell surface proteins.
Chen I, Choi YA, Ting AY. Chen I, et al. J Am Chem Soc. 2007 May 23;129(20):6619-25. doi: 10.1021/ja071013g. Epub 2007 May 2. J Am Chem Soc. 2007. PMID: 17472384 Free PMC article. - In vivo biotinylation of bacterial magnetic particles by a truncated form of Escherichia coli biotin ligase and biotin acceptor peptide.
Maeda Y, Yoshino T, Matsunaga T. Maeda Y, et al. Appl Environ Microbiol. 2010 Sep;76(17):5785-90. doi: 10.1128/AEM.00916-10. Epub 2010 Jul 9. Appl Environ Microbiol. 2010. PMID: 20622127 Free PMC article. - Molecular biology of biotin attachment to proteins.
Chapman-Smith A, Cronan JE Jr. Chapman-Smith A, et al. J Nutr. 1999 Feb;129(2S Suppl):477S-484S. doi: 10.1093/jn/129.2.477S. J Nutr. 1999. PMID: 10064313 Review. - Microbial biotin protein ligases aid in understanding holocarboxylase synthetase deficiency.
Pendini NR, Bailey LM, Booker GW, Wilce MC, Wallace JC, Polyak SW. Pendini NR, et al. Biochim Biophys Acta. 2008 Jul-Aug;1784(7-8):973-82. doi: 10.1016/j.bbapap.2008.03.011. Epub 2008 Apr 9. Biochim Biophys Acta. 2008. PMID: 18442489 Review.
Cited by
- Choosing the Probe for Single-Molecule Fluorescence Microscopy.
Schirripa Spagnolo C, Luin S. Schirripa Spagnolo C, et al. Int J Mol Sci. 2022 Nov 29;23(23):14949. doi: 10.3390/ijms232314949. Int J Mol Sci. 2022. PMID: 36499276 Free PMC article. Review. - Single cell fluorescence imaging using metal plasmon-coupled probe.
Zhang J, Fu Y, Lakowicz JR. Zhang J, et al. Bioconjug Chem. 2007 May-Jun;18(3):800-5. doi: 10.1021/bc0603384. Epub 2007 Mar 22. Bioconjug Chem. 2007. PMID: 17375898 Free PMC article. - Crystallization and preliminary X-ray crystallographic studies of the biotin carboxyl carrier protein and biotin protein ligase complex from Pyrococcus horikoshii OT3.
Bagautdinov B, Matsuura Y, Bagautdinova S, Kunishima N. Bagautdinov B, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007 Apr 1;63(Pt 4):334-7. doi: 10.1107/S1744309107011967. Epub 2007 Mar 30. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007. PMID: 17401210 Free PMC article. - Transglutaminase-catalyzed site-specific conjugation of small-molecule probes to proteins in vitro and on the surface of living cells.
Lin CW, Ting AY. Lin CW, et al. J Am Chem Soc. 2006 Apr 12;128(14):4542-3. doi: 10.1021/ja0604111. J Am Chem Soc. 2006. PMID: 16594669 Free PMC article. - Protein-assisted self-assembly of multifunctional nanoparticles.
Nikitin MP, Zdobnova TA, Lukash SV, Stremovskiy OA, Deyev SM. Nikitin MP, et al. Proc Natl Acad Sci U S A. 2010 Mar 30;107(13):5827-32. doi: 10.1073/pnas.1001142107. Epub 2010 Mar 15. Proc Natl Acad Sci U S A. 2010. PMID: 20231484 Free PMC article.
References
- Gao, X., Yang, L., Petros, J. A., Marshall, F. F., Simons, J. W. & Nie, S. (2005) Curr. Opin. Biotechnol. 16, 63–72. - PubMed
- Dahan, M., Levi, S., Luccardini, C., Rostaing, P., Riveau, B. & Triller, A. (2003) Science 302, 442–445. - PubMed
- Chan, W. C. & Nie, S. (1998) Science 281, 2016–2018. - PubMed
- Bruchez, M., Jr., Moronne, M., Gin, P., Weiss, S. & Alivisatos, A. P. (1998) Science 281, 2013–2016. - PubMed
- Han, M., Gao, X., Su, J. Z. & Nie, S. (2001) Nat. Biotechnol. 19, 631–635. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous