Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud - PubMed (original) (raw)
Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud
Catherine Olivier et al. Mol Cell Biol. 2005 Jun.
Abstract
In Saccharomyces cerevisiae, over twenty mRNAs localize to the bud tip of daughter cells, playing roles in processes as different as mating type switching and plasma membrane targeting. The localization of these transcripts depends on interactions between a cis-acting localization element(s) or zipcodes and the RNA-binding protein She2p. While previous studies identified four different localization elements in the bud-localized ASH1 mRNA, the main determinants for She2p recognition are still unknown. To investigate the RNA-binding specificity of She2p, we isolated She2p-binding RNAs by in vivo selection from libraries of partially randomized ASH1 localization elements. The RNAs isolated contained a similar loop-stem-loop structure with a highly conserved CGA triplet in one loop and a single conserved cytosine in the other loop. Mutating these conserved nucleotides or the stem separating them resulted in the loss of She2p binding and in the delocalization of a reporter mRNA. Using this information, we identified the same RNA motif in two other known bud-localized transcripts, suggesting that this motif is conserved among bud-localized mRNAs. These results show that mRNAs with zipcodes lacking primary sequence similarity can rely on a few conserved nucleotides properly oriented in their three-dimensional structure in order to be recognized by the same localization machinery.
Figures
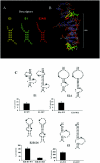
FIG.1.
Four ASH1 localization elements interact with She2p in a yeast three-hybrid assay. (A) Predicted secondary structures of the ASH1 localization elements E1, E2A, E2B, and E3. Secondary structures are described with latin numbered stems (S) and loops (L). For the element E2B, two separate domains have been previously identified (8), but only the secondary structure of domain 1 has been defined. (B) The yeast strain YBZ1 expresses a dimerized bacteriophage MS2 RNA-binding protein fused to a single LexA DNA-binding domain (MS2-LexA), with either wild-type (capital letters) or knockout (lowercase) SHE2 gene. These strains were transformed with a plasmid expressing the GAL4 activation domain fused with the C-terminal domain of She3p (She3-Cterm-AD), along with a plasmid expressing an hybrid RNA containing the indicated RNA fused to two MS2 binding sites. The formation of a She2p-hybrid RNA complex allows the recruitment of the She3-Cterm-AD and MS2-LexA proteins and the activation of the reporter genes. Controls: empty vector (pIIIA/MS2-2) and the iron response element RNA (IRE).
FIG.2.
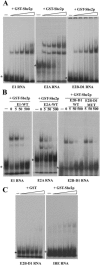
Recombinant GST-She2p binds to the ASH1 localization element RNAs in vitro. (A) Electrophoretic mobility shift assays (EMSA) of GST-She2p with the localization elements RNA E1, E2A, and E2B-D1. Increasing concentration of recombinant GST-She2p (0, 0.5, 0.9, 1.8, 3.7, and 7.4 μM) was added to 32P-labeled localization element RNA. The free RNA is indicated by an asterisk. (B) Competition of the RNA-protein complex with unlabeled localization element RNA E1, E2A and E2B-D1. A constant quantity of GST-She2p (3.7 μM for E1 and E2B-D1, 0.9 μM for E2A) was combined to an increasing concentration of unlabeled RNA (5×, 50×, and 500×). The mutant M16 of the element E2B-D1 was used as a competitor (E2D-D1 MUT). (C) Controls using GST or the IRE RNA. For GST, increasing concentration of recombinant GST (0, 0.5, 0.9, 1.8, 3.7, and 7.4 μM) was added to the 32P-labeled localization element E2B-D1 RNA. For the IRE RNA, increasing concentration of recombinant GST-She2p (0, 0.5, 0.9, 1.8, 3.7, and 7.4 μM) was added to the 32P-labeled IRE RNA.
FIG. 3.
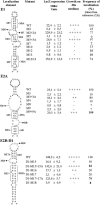
Identification of the She2p-binding domain in the localization elements E1, E2A, and E2B-D1. Mutated RNAs were fused to the MS2 binding RNA in the pIIIA/MS2-2 plasmid and transformed in yeasts YBZ1 containing the plasmid pGAD-She3-Ct. The regions mutated are indicated by brackets (for stems) or in gray letters (for loops). The strength of the interaction of the RNA mutants with the She3p-Ct/She2p complex in the yeast three-hybrid assay is reported as lacZ expression level (Miller units) or growth on a plate lacking histidine. Values for the percentage of localization of the mutants are from references and .
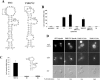
FIG. 4.
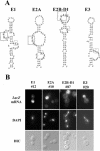
In vivo selection of partially randomized ASH1 localization element RNAs. (A) Diagram of the mutagenesis performed on the She2p-binding domains of the four ASH1 localization elements. Partially randomized positions are underlined. Nucleotides conserved in >95% of the clones isolated are indicated in capital letters and italicized. The conserved CGA sequence is boxed with large dotted lines, whereas the conserved cytosine in E1, E2A, and E2B-D1 is boxed with small dotted lines. A cytosine at an analogous position on the element E3 is indicated by an asterisk. (B) Fluorescent in situ hybridization on the lacZ mRNA fused to ASH1 localization element variants isolated from the three-hybrid screen. Numbers correspond to clones listed in the Table 1. DAPI, DNA staining; DIC, Nomarski.
FIG. 5.
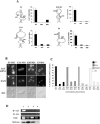
Two conserved cytosines in the four ASH1 localization elements define a She2p-binding motif. (A) Mutagenesis of each highly conserved cytosine to uracil or adenine in the ASH1 localization element results in the loss of interaction with She2p in the yeast three-hybrid assay. Mutated cytosines are boxed. lacZ expression is in Miller units. (B) Fluorescent in situ hybridization on the lacZ mRNA fused to ASH1 localizationelements mutated on the conserved cytosines. The presence of these mutations in the localization elements leads to the delocalization of the lacZ-E1, lacZ-E2A, lacZ-E2B, and lacZ-E3 reporter mRNAs. DAPI, nuclear DNA; DIC, Nomarski. (C) Measurement of the percentage of budding yeast cells with bud-localized lacZ mRNA fused to wild-type or mutated localization elements. (D) Coimmunoprecipitation of She2p-Myc and lacZ-E1 mRNAs was disrupted by mutation of the conserved cytosines in the element E1. Reverse transcription-PCR detection of the lacZ mRNA fused to wild-type localization element E1 (lane 1) or mutants M15 (lane 2) and M16 (lane 3) of the element E1 was performed on either total yeast extract (total) or on the pellet of the immunoprecipitated Myc-tagged She2p (IP+RT). A yeast strain expressing the lacZ-E1 mRNA and She2p without a Myc tag (lane 4) or reverse transcription-PCRs on the immunoprecipitates without reverse transcriptase (IP-RT) were used as controls.
FIG. 6.
Distance between the two conserved cytosines is important for She2p recognition. (A) Descriptors used in the program MC-SEARCH. (B) Overlap of three structures, one from each descriptor, that were identified by the program. The cytosines are colored according to their descriptor motif in panel A. The distance indicated corresponds to the distance between the 3′ phosphates of the two cytosines. For E3 (yellow), the motif comes from the 23S rRNA, starting at position 295 (PDB 1JZY). For E1 (green), the motif comes from an hairpin similar to the P5Abc region of the group I intron, starting at position 4 (PDB 1EOR). For E2A/B (red), the motif comes from the 23S rRNA, starting at the position 1463 (PDB 1M1K). The root mean square deviation in their common region (the stem structure) is 1.9 Å. (C) Increasing the length of the stem separating the two cytosines in each localization element leads to a decreased interaction with She2p in the yeast three-hybrid assay. The conserved cytosines are labeled in gray. lacZ expression is in Miller units.
FIG. 7.
Identification of a She2p-binding motif in the bud-localized IST2 and YMR171C mRNAs. (A) Predicted secondary structures of the IST2 and YMR171C mRNA localization elements. The conserved CGA sequence is boxed with plain lines, whereas the conserved cytosine is boxed with dotted lines. The nucleotides are numbered starting from the adenosine of the start codon as +1. (B) Three-hybrid assays using yeast strain YBZ1 with either wild-type or knockout SHE2 gene, transformed with a plasmid expressing the GAL4 activation domain fused with the C-terminal domain of She3p, along with a plasmid expressing one of the indicated MS2 fusion RNAs. Controls: empty vector (pIIIA/MS2-2) and the iron response element RNA (IRE). (C) Mutagenesis of each highly conserved cytosine in the YMR171c mRNA localization element results in the loss of interaction with She2p in the yeast three-hybrid assay. (D) Fluorescent in situ hybridization on the lacZ mRNA fused to the IST2 and wild-type, or mutated, YMR171C mRNA localization elements. The percentage of budding yeast cells with bud-localized lacZ mRNA is indicated. DAPI, nuclear DNA; DIC, Nomarski.
Similar articles
- She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p.
Böhl F, Kruse C, Frank A, Ferring D, Jansen RP. Böhl F, et al. EMBO J. 2000 Oct 16;19(20):5514-24. doi: 10.1093/emboj/19.20.5514. EMBO J. 2000. PMID: 11032818 Free PMC article. - Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud.
Shen Z, St-Denis A, Chartrand P. Shen Z, et al. Genes Dev. 2010 Sep 1;24(17):1914-26. doi: 10.1101/gad.1937510. Epub 2010 Aug 16. Genes Dev. 2010. PMID: 20713510 Free PMC article. - Association of the yeast RNA-binding protein She2p with the tubular endoplasmic reticulum depends on membrane curvature.
Genz C, Fundakowski J, Hermesh O, Schmid M, Jansen RP. Genz C, et al. J Biol Chem. 2013 Nov 8;288(45):32384-32393. doi: 10.1074/jbc.M113.486431. Epub 2013 Sep 20. J Biol Chem. 2013. PMID: 24056370 Free PMC article. - RNA asymmetric distribution and daughter/mother differentiation in yeast.
Darzacq X, Powrie E, Gu W, Singer RH, Zenklusen D. Darzacq X, et al. Curr Opin Microbiol. 2003 Dec;6(6):614-20. doi: 10.1016/j.mib.2003.10.005. Curr Opin Microbiol. 2003. PMID: 14662358 Free PMC article. Review. - RNA localization in yeast: moving towards a mechanism.
Gonsalvez GB, Urbinati CR, Long RM. Gonsalvez GB, et al. Biol Cell. 2005 Jan;97(1):75-86. doi: 10.1042/BC20040066. Biol Cell. 2005. PMID: 15601259 Review.
Cited by
- Statistical evidence for conserved, local secondary structure in the coding regions of eukaryotic mRNAs and pre-mRNAs.
Meyer IM, Miklós I. Meyer IM, et al. Nucleic Acids Res. 2005 Nov 7;33(19):6338-48. doi: 10.1093/nar/gki923. Print 2005. Nucleic Acids Res. 2005. PMID: 16275783 Free PMC article. - Cis-acting determinants of asymmetric, cytoplasmic RNA transport.
Jambhekar A, Derisi JL. Jambhekar A, et al. RNA. 2007 May;13(5):625-42. doi: 10.1261/rna.262607. RNA. 2007. PMID: 17449729 Free PMC article. Review. - COMIT: identification of noncoding motifs under selection in coding sequences.
Kural D, Ding Y, Wu J, Korpi AM, Chuang JH. Kural D, et al. Genome Biol. 2009;10(11):R133. doi: 10.1186/gb-2009-10-11-r133. Epub 2009 Nov 20. Genome Biol. 2009. PMID: 19930548 Free PMC article. - An RNA transport system in Candida albicans regulates hyphal morphology and invasive growth.
Elson SL, Noble SM, Solis NV, Filler SG, Johnson AD. Elson SL, et al. PLoS Genet. 2009 Sep;5(9):e1000664. doi: 10.1371/journal.pgen.1000664. Epub 2009 Sep 25. PLoS Genet. 2009. PMID: 19779551 Free PMC article. - Cotranscriptional assembly of mRNP complexes that determine the cytoplasmic fate of mRNA.
Forget A, Chartrand P. Forget A, et al. Transcription. 2011 Mar;2(2):86-90. doi: 10.4161/trns.2.2.14857. Transcription. 2011. PMID: 21468235 Free PMC article.
References
- Addess, K. J., J. P. Basilion, R. D. Klausner, T. A. Rouault, and A. Pardi. 1997. Structure and dynamics of the iron responsive element RNA: implications for binding of the RNA by iron regulatory binding proteins1. J. Mol. Biol. 274:72-83. - PubMed
- Bashirullah, A., R. L. Cooperstock, and H. D. Lipshitz. 1998. RNA localization in development. Annu. Rev. Biochem. 67:335-394. - PubMed
- Bertrand, E., P. Chartrand, M. Schaefer, S. M. Shenoy, R. H. Singer, and R. M. Long. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 2:437-445. - PubMed
- Bullock, S. L., and D. Ish-Horowicz. 2001. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 414:611-616. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials