Developmentally induced changes in transcriptional program alter spatial organization across chromosomes - PubMed (original) (raw)
Developmentally induced changes in transcriptional program alter spatial organization across chromosomes
Jason M Casolari et al. Genes Dev. 2005.
Abstract
Although the spatial location of genes within the nucleus has been implicated in their transcriptional status, little is known about the dynamics of gene location that accompany large-scale changes in gene expression. The mating of haploid yeast Saccharomyces cerevisiae is accompanied by a large-scale change in transcription and developmental program. We examined changes in nuclear organization that accompany stimulus by the mating pheromone alpha factor and found that most alpha-factor-induced genes become associated with components of the nuclear envelope. The myosin-like protein Mlp1, which has been implicated in mRNA export, was further shown to exhibit RNA dependence in its association with alpha-factor-induced genes. High-resolution mapping of association of chromosome III with Mlp1 revealed alpha-factor-dependent determinants of nuclear pore association, including origins of replication, specific intergenic regions, and the 3' ends of transcriptionally activated genes. Taken together, these results reveal RNA- and DNA-dependent determinants of nuclear organization as well as a detailed picture of how an entire chromosome alters its spatial conformation in response to a developmental cue.
Figures
Figure 1.
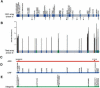
Correlation among genome occupancy, transcriptional activation, and peripheral localization. All genes in the genomic occupancy profiles of Mlp1 and RanGEF/Prp20 were assigned a percentile rank ranging from 0% (unbound) to 100% (highest binding). The distributions of the 49 genes induced threefold or more upon α-factor treatment (Spellman et al. 1998) within the Mlp1 (A) and RanGEF/Prp20 (B) data sets are shown for untreated cells (white bars) and cells treated with 30 nM α factor for 2 h (black bars). A bin size of 10% was used. Combined FISH/IF of the FIG2 locus in cells that were untreated (C) and stimulated with 30 nM α factor for 2 h (D). The FIG2 probe is visualized as a single green spot (see yellow arrow) with nuclear pore staining in red. The outline of each stimulated cell is shown in white, with the shmoo projection highlighted by a white arrow. (E) Mlp1 genomic location analysis was carried out as in A except that cells were fixed for only 5 min plus or minus RNase prior to immunoprecipitation (F). The distribution of the 49 genes induced three-fold or more upon α-factor treatment are shown for untreated cells (white bars) and cells treated with 30 nM α factor for 2 h (black bars).
Figure 2.
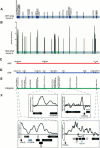
Mlp1 occupancy on chromosome III using whole-genome ORF and high-resolution tiled arrays. (A) Genes bound by Mlp1 (IP/WCE > 1.0 and _p_-value ≤ 0.05) on whole-genome ORF arrays (Casolari et al. 2004) are displayed (blue) on a spatial representation of S. cerevisiae chromosome III (gray bar). (B) High-resolution mapping of Mlp1-bound probes (see Materials and Methods) on tiled arrays is shown on a representation of chromosome III (gray bar). Data having a standard deviation >2.0 and a moving average >2.0 are shown. Binding to probes within ARS elements (red), ORFs (blue), and intergenic sequences (green) are depicted with colored bars on a spatial representation of the chromosome. (C_–_E) Significantly bound probes were mapped to the corresponding ARSes, ORFs, and intergenic sequences and are labeled on representations of chromosome III. (F) Mlp1 binding to regions of chromosome III is displayed with a moving average of nine adjacent probes. Black arrows indicate the direction of transcription for each ORF. Gray arrows represent ORFs considered dubious by the Saccharomyces Genome Database, and open boxes represent transposon sequences.
Figure 3.
Mlp1 occupancy on chromosome III after stimulation with α factor. Genes bound by Mlp1 on whole-genome ORF arrays (A) and tiled arrays (B) after stimulation with α factor are displayed on a spatial representation of S. cerevisiae chromosome III (gray bar). α-Factor-induced changes in Mlp1 binding to ORF arrays, relative to untreated cells, are represented with upward (gain in binding) and downward (loss of binding) arrows. (C_–_E) Binding to ARS, ORF, and intergenic regions are displayed as in Figure 2.
Figure 4.
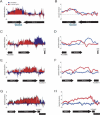
Local changes in Mlp1 occupancy upon α-factor stimulation of transcription. Mlp1 binding to transcriptionally active regions are shown. Standard deviations for individual probes (A,C,E,G) as well as a moving average of binding for nine adjacent probes (B,D,F,H) are shown. ORFs are displayed as in Figure 2.
Figure 5.
Global changes in Mlp1 occupancy reflect conformational changes in chromosome III relative to the nuclear periphery. (A) Mlp1 binding on chromosome III in untreated and α-factor-stimulated cells is shown. Binding to sites separated by <8.5 kb is considered semicontinuous and represented with a horizontal line at the nuclear periphery. Binding gaps of >8.5 kb are represented by a loop extending into the nuclear interior that is proportional to the distance between Mlp1-binding sites. Regions where data are unavailable are represented by breaks in the line. (B) The regions of binding that are held constant under the two conditions, stretching from 0 to ∼28 kb (region 1), ∼120 to 180 kb (region 3), and ∼285 to 300 kb (region 5), are displayed in black. Regions exhibiting pronounced changes in binding pattern between the two conditions (regions 2 and 4) are displayed in red.
Similar articles
- Nuclear pore association confers optimal expression levels for an inducible yeast gene.
Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Taddei A, et al. Nature. 2006 Jun 8;441(7094):774-8. doi: 10.1038/nature04845. Nature. 2006. PMID: 16760983 - Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression.
Dieppois G, Stutz F. Dieppois G, et al. J Cell Sci. 2010 Jun 15;123(Pt 12):1989-99. doi: 10.1242/jcs.053694. J Cell Sci. 2010. PMID: 20519581 - SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope.
Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, Nehrbass U. Cabal GG, et al. Nature. 2006 Jun 8;441(7094):770-3. doi: 10.1038/nature04752. Nature. 2006. PMID: 16760982 - Post-transcriptional regulation of gene expression in yeast under ethanol stress.
Izawa S, Inoue Y. Izawa S, et al. Biotechnol Appl Biochem. 2009 May 6;53(Pt 2):93-9. doi: 10.1042/BA20090036. Biotechnol Appl Biochem. 2009. PMID: 19397495 Review. - The nuclear envelope and transcriptional control.
Akhtar A, Gasser SM. Akhtar A, et al. Nat Rev Genet. 2007 Jul;8(7):507-17. doi: 10.1038/nrg2122. Epub 2007 Jun 5. Nat Rev Genet. 2007. PMID: 17549064 Review.
Cited by
- Dynamic association of NUP98 with the human genome.
Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Liang Y, et al. PLoS Genet. 2013;9(2):e1003308. doi: 10.1371/journal.pgen.1003308. Epub 2013 Feb 28. PLoS Genet. 2013. PMID: 23468646 Free PMC article. - Subnuclear positioning and interchromosomal clustering of the GAL1-10 locus are controlled by separable, interdependent mechanisms.
Brickner DG, Sood V, Tutucci E, Coukos R, Viets K, Singer RH, Brickner JH. Brickner DG, et al. Mol Biol Cell. 2016 Oct 1;27(19):2980-93. doi: 10.1091/mbc.E16-03-0174. Epub 2016 Aug 3. Mol Biol Cell. 2016. PMID: 27489341 Free PMC article. - One, Two, Three: Polycomb Proteins Hit All Dimensions of Gene Regulation.
Del Prete S, Mikulski P, Schubert D, Gaudin V. Del Prete S, et al. Genes (Basel). 2015 Jul 10;6(3):520-42. doi: 10.3390/genes6030520. Genes (Basel). 2015. PMID: 26184319 Free PMC article. Review. - Coiled coil structures and transcription: an analysis of the S. cerevisiae coilome.
Barbara KE, Willis KA, Haley TM, Deminoff SJ, Santangelo GM. Barbara KE, et al. Mol Genet Genomics. 2007 Aug;278(2):135-47. doi: 10.1007/s00438-007-0237-x. Epub 2007 May 3. Mol Genet Genomics. 2007. PMID: 17476531 - Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes.
Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Capelson M, et al. Cell. 2010 Feb 5;140(3):372-83. doi: 10.1016/j.cell.2009.12.054. Cell. 2010. PMID: 20144761 Free PMC article.
References
- Aebi M., Clark, M.W., Vijayraghavan, U., and Abelson, J. 1990. A yeast mutant, PRP20, altered in mRNA metabolism and maintenance of the nuclear structure, is defective in a gene homologous to the human gene RCC1 which is involved in the control of chromosome condensation. Mol. Gen. Genet. 224: 72–80. - PubMed
- Andrulis E.D., Neiman, A.M., Zappulla, D.C., and Sternglanz, R. 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394: 592–595. - PubMed
- Azuma Y. and Dasso, M. 2000. The role of Ran in nuclear function. Curr. Opin. Cell Biol. 12: 302–307. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases