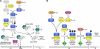Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation - PubMed (original) (raw)
Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation
Cristina Pina et al. Plant Physiol. 2005 Jun.
Abstract
Upon germination, pollen forms a tube that elongates dramatically through female tissues to reach and fertilize ovules. While essential for the life cycle of higher plants, the genetic basis underlying most of the process is not well understood. We previously used a combination of flow cytometry sorting of viable hydrated pollen grains and GeneChip array analysis of one-third of the Arabidopsis (Arabidopsis thaliana) genome to define a first overview of the pollen transcriptome. We now extend that study to approximately 80% of the genome of Arabidopsis by using Affymetrix Arabidopsis ATH1 arrays and perform comparative analysis of gene family and gene ontology representation in the transcriptome of pollen and vegetative tissues. Pollen grains have a smaller and overall unique transcriptome (6,587 genes expressed) with greater proportions of selectively expressed (11%) and enriched (26%) genes than any vegetative tissue. Relative gene ontology category representations in pollen and vegetative tissues reveal a functional skew of the pollen transcriptome toward signaling, vesicle transport, and the cytoskeleton, suggestive of a commitment to germination and tube growth. Cell cycle analysis reveals an accumulation of G2/M-associated factors that may play a role in the first mitotic division of the zygote. Despite the relative underrepresentation of transcription-associated transcripts, nonclassical MADS box genes emerge as a class with putative unique roles in pollen. The singularity of gene expression control in mature pollen grains is further highlighted by the apparent absence of small RNA pathway components.
Figures
Figure 1.
Snail view representation and principal component analysis of tissue-dependent gene expression patterns. A, 18,321 genes used for a snail view representation were ranked counterclockwise from top according to decreasing mean expression in seedlings. For each tissue, the mean pattern in seedlings (black line) is coplotted with the mean pattern of the specific tissue (gray dots). The radius encodes the logarithm of gene expression (values <1 were set to 1 for better visualization). B, Two thousand of the 3,734 genes expressed in the vegetative tissues and in pollen were analyzed by projecting the 5 tissue types (replicate datasets are shown) on the first 2 principal components. The first principal component separates pollen from the vegetative tissues, while the second principal component shows a further separation of the vegetative tissues within themselves.
Figure 2.
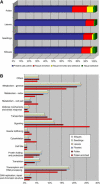
Proportions of enriched and selectively expressed genes and classification of biological activities in Arabidopsis tissues. A, Genes called present in each of the Arabidopsis tissues were analyzed in terms of enriched and/or selective expression. Tissue-enriched genes have expression levels in any particular tissue that are at least 1.2-fold higher than in all the remaining tissues; selectively expressed genes are those called present only in the particular tissue analyzed. B, 8,463 genes represented on the ATH1 GeneChip that were classified into at least one GO category (as of September 2003) were regrouped into 14 biological activity classes. The transcription and RNA-processing classification includes transcription factors, basic transcriptional machinery, and RNA-processing factors; the cell fate category covers cell cycle, cell differentiation, and apoptosis-related molecules; transporters refers to membrane channels. The proportion of the genes expressed in each tissue assigned to the different activity classes is represented. The separated bars show this classification for the pollen-enriched genes.
Figure 3.
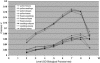
Plot of intertissue distances for the different levels of the GO tree of biological process terms. Overall differences in term frequencies between any two Arabidopsis tissues at each level of the GO biological process tree were calculated using Cramer's V and interpreted as a measure of the distance or dissimilarity between the two tissues. Results vary between 0 and 1, lower values reflecting more similar relative frequencies of terms. A total of 16,024 genes represented on the ATH1 GeneChip and classified into GO categories (as of December 2003) were included in the analysis. The plot is representative of results obtained for molecular function and component GO trees.
Figure 4.
Overview of potential cell cycle regulation in pollen. Pollen gene expression data for core genes of the cell cycle (see also Table II) are overlaid on a schematic overview of the G1/S (A) and G2/M (B) transitions. For proteins colored light gray, no transcripts were detected in pollen (absent call). Transcripts expressed in pollen (present call) are either expressed at a comparable level to the vegetative tissues or enriched (arrow up; fold change value) or depleted (arrow down; fold change value). Question marks indicate pathways that are still not fully demonstrated experimentally. (Modified from De Veylder et al., 2003).
Similar articles
- Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome.
Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA. Becker JD, et al. Plant Physiol. 2003 Oct;133(2):713-25. doi: 10.1104/pp.103.028241. Epub 2003 Sep 18. Plant Physiol. 2003. PMID: 14500793 Free PMC article. - Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa.
Wei LQ, Xu WY, Deng ZY, Su Z, Xue Y, Wang T. Wei LQ, et al. BMC Genomics. 2010 May 28;11:338. doi: 10.1186/1471-2164-11-338. BMC Genomics. 2010. PMID: 20507633 Free PMC article. - Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis.
Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH. Wang Y, et al. Plant Physiol. 2008 Nov;148(3):1201-11. doi: 10.1104/pp.108.126375. Epub 2008 Sep 5. Plant Physiol. 2008. PMID: 18775970 Free PMC article. - Unveiling the gene-expression profile of pollen.
da Costa-Nunes JA, Grossniklaus U. da Costa-Nunes JA, et al. Genome Biol. 2003;5(1):205. doi: 10.1186/gb-2003-5-1-205. Epub 2003 Dec 24. Genome Biol. 2003. PMID: 14709169 Free PMC article. Review. - Of weeds and men: what genomes teach us about plant cell biology.
Assaad FF. Assaad FF. Curr Opin Plant Biol. 2001 Dec;4(6):478-87. doi: 10.1016/s1369-5266(00)00204-1. Curr Opin Plant Biol. 2001. PMID: 11641062 Review.
Cited by
- Pollen transcriptome analysis of Solanum tuberosum (2n = 4x = 48), S. demissum (2n = 6x = 72), and their reciprocal F1 hybrids.
Sanetomo R, Hosaka K. Sanetomo R, et al. Plant Cell Rep. 2013 May;32(5):623-36. doi: 10.1007/s00299-013-1395-4. Epub 2013 Feb 22. Plant Cell Rep. 2013. PMID: 23430172 - Intracellular auxin transport in pollen: PIN8, PIN5 and PILS5.
Dal Bosco C, Dovzhenko A, Palme K. Dal Bosco C, et al. Plant Signal Behav. 2012 Nov;7(11):1504-5. doi: 10.4161/psb.21953. Epub 2012 Sep 18. Plant Signal Behav. 2012. PMID: 22990451 Free PMC article. - Bioinformatics resources for pollen.
Ambrosino L, Bostan H, Ruggieri V, Chiusano ML. Ambrosino L, et al. Plant Reprod. 2016 Jun;29(1-2):133-47. doi: 10.1007/s00497-016-0284-8. Epub 2016 Jun 8. Plant Reprod. 2016. PMID: 27271281 Review. - Pollen proteomics: from stress physiology to developmental priming.
Chaturvedi P, Ghatak A, Weckwerth W. Chaturvedi P, et al. Plant Reprod. 2016 Jun;29(1-2):119-32. doi: 10.1007/s00497-016-0283-9. Epub 2016 Jun 8. Plant Reprod. 2016. PMID: 27271282 Free PMC article. Review. - Gene, protein, and network of male sterility in rice.
Wang K, Peng X, Ji Y, Yang P, Zhu Y, Li S. Wang K, et al. Front Plant Sci. 2013 Apr 11;4:92. doi: 10.3389/fpls.2013.00092. eCollection 2013. Front Plant Sci. 2013. PMID: 23596452 Free PMC article.
References
- Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, Vergara-Silva F, Yanofsky MF (2000. a) MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J 24: 457–466 - PubMed
- Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363 - PubMed
- Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301: 336–338 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases