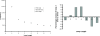Prevalence of quadruplexes in the human genome - PubMed (original) (raw)
Prevalence of quadruplexes in the human genome
Julian L Huppert et al. Nucleic Acids Res. 2005.
Abstract
Guanine-rich DNA sequences of a particular form have the ability to fold into four-stranded structures called G-quadruplexes. In this paper, we present a working rule to predict which primary sequences can form this structure, and describe a search algorithm to identify such sequences in genomic DNA. We count the number of quadruplexes found in the human genome and compare that with the figure predicted by modelling DNA as a Bernoulli stream or as a Markov chain, using windows of various sizes. We demonstrate that the distribution of loop lengths is significantly different from what would be expected in a random case, providing an indication of the number of potentially relevant quadruplex-forming sequences. In particular, we show that there is a significant repression of quadruplexes in the coding strand of exonic regions, which suggests that quadruplex-forming patterns are disfavoured in sequences that will form RNA.
Figures
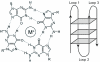
Figure 1
Left: hydrogen bond pattern in a G-tetrad. A monvalent cation occupies the central position. Right: Schematic diagram of a unimolecular G-quadruplex structure.
Figure 2
Process for generating Markov windowed simulates. A real chromosome (top) is separated into discrete windows. For each of these, a table of base and diad frequencies is generated (middle), which is then used to generate a simulated window (bottom), which are then joined to produce the replicate chromosome.
Figure 3
Left: frequency distributions of loops of lengths 1–7 bases for the entire human genome. Right: percentage excesses of loop 2 counts over the averages of loops 1 and 3 for the entire human genome.
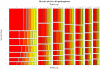
Figure 4
Mosaic plot representing the loop lengths of all putative quadruplexes found in the human genome. The seven principle columns represent the lengths of the first loop, the seven rows the lengths of the second loop, and the seven segments in each box the lengths of the third loop. The area of each box is proportional to the number of sequences found with that combination of loop lengths. The plot was produced using the program R, (
) using the command mosaicplot.
Similar articles
- QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences.
Kikin O, D'Antonio L, Bagga PS. Kikin O, et al. Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W676-82. doi: 10.1093/nar/gkl253. Nucleic Acids Res. 2006. PMID: 16845096 Free PMC article. - Highly prevalent putative quadruplex sequence motifs in human DNA.
Todd AK, Johnston M, Neidle S. Todd AK, et al. Nucleic Acids Res. 2005 May 24;33(9):2901-7. doi: 10.1093/nar/gki553. Print 2005. Nucleic Acids Res. 2005. PMID: 15914666 Free PMC article. - G-quadruplexes in promoters throughout the human genome.
Huppert JL, Balasubramanian S. Huppert JL, et al. Nucleic Acids Res. 2007;35(2):406-13. doi: 10.1093/nar/gkl1057. Epub 2006 Dec 14. Nucleic Acids Res. 2007. PMID: 17169996 Free PMC article. - NMR spectroscopy of G-quadruplexes.
Adrian M, Heddi B, Phan AT. Adrian M, et al. Methods. 2012 May;57(1):11-24. doi: 10.1016/j.ymeth.2012.05.003. Epub 2012 May 24. Methods. 2012. PMID: 22633887 Review. - A guide to computational methods for G-quadruplex prediction.
Puig Lombardi E, Londoño-Vallejo A. Puig Lombardi E, et al. Nucleic Acids Res. 2020 Jan 10;48(1):1-15. doi: 10.1093/nar/gkz1097. Nucleic Acids Res. 2020. PMID: 31754698 Free PMC article. Review.
Cited by
- Zika Virus Genomic RNA Possesses Conserved G-Quadruplexes Characteristic of the Flaviviridae Family.
Fleming AM, Ding Y, Alenko A, Burrows CJ. Fleming AM, et al. ACS Infect Dis. 2016 Oct 14;2(10):674-681. doi: 10.1021/acsinfecdis.6b00109. Epub 2016 Aug 12. ACS Infect Dis. 2016. PMID: 27737553 Free PMC article. - Aptamer selection based on G4-forming promoter region.
Yoshida W, Saito T, Yokoyama T, Ferri S, Ikebukuro K. Yoshida W, et al. PLoS One. 2013 Jun 4;8(6):e65497. doi: 10.1371/journal.pone.0065497. Print 2013. PLoS One. 2013. PMID: 23750264 Free PMC article. - Diversity of Parallel Guanine Quadruplexes Induced by Guanine Substitutions.
Bednářová K, Vorlíčková M, Renčiuk D. Bednářová K, et al. Int J Mol Sci. 2020 Aug 25;21(17):6123. doi: 10.3390/ijms21176123. Int J Mol Sci. 2020. PMID: 32854410 Free PMC article. - Replication of Structured DNA and its implication in epigenetic stability.
Cea V, Cipolla L, Sabbioneda S. Cea V, et al. Front Genet. 2015 Jun 16;6:209. doi: 10.3389/fgene.2015.00209. eCollection 2015. Front Genet. 2015. PMID: 26136769 Free PMC article. Review. - Getting Ready for the Dance: FANCJ Irons Out DNA Wrinkles.
Bharti SK, Awate S, Banerjee T, Brosh RM. Bharti SK, et al. Genes (Basel). 2016 Jul 1;7(7):31. doi: 10.3390/genes7070031. Genes (Basel). 2016. PMID: 27376332 Free PMC article. Review.
References
- Guschlbauer W., Chantot J.F., Theile D. Four-stranded nucleic structures 25 years later: from guanosine gels to telomere DNA. J. Biomol. Struct. Dyn. 1990;8:491–511. - PubMed
- Blackburn E.H. Telomeres and their synthesis. Science. 1990;249:489–490. - PubMed
- Blackburn E.H. Structure and function of telomeres. Nature. 1991;350:569–573. - PubMed
- Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources