RNA interference: more than a research tool in the vertebrates' adaptive immunity - PubMed (original) (raw)
RNA interference: more than a research tool in the vertebrates' adaptive immunity
Johnson Mak. Retrovirology. 2005.
Abstract
In recent years, RNA silencing, usage of small double stranded RNAs of approximately 21 - 25 base pairs to regulate gene expression, has emerged as a powerful research tool to dissect the role of unknown host cell factors in this 'post-genomic' era. While the molecular mechanism of RNA silencing has not been precisely defined, the revelation that small RNA molecules are equipped with this regulatory function has transformed our thinking on the role of RNA in many facets of biology, illustrating the complexity and the dynamic interplay of cellular regulation. As plants and invertebrates lack the protein-based adaptive immunity that are found in jawed vertebrates, the ability of RNA silencing to shut down gene expression in a sequence-specific manner offers an explanation of how these organisms counteract pathogen invasions into host cells. It has been proposed that this type of RNA-mediated defence mechanism is an ancient form of immunity to offset the transgene-, transposon- and virus-mediated attack. However, whether 1) RNA silencing is a natural immune response in vertebrates to suppress pathogen invasion; or 2) vertebrate cells have evolved to counteract invasion in a 'RNA silencing' independent manner remains to be determined. A number of recent reports have provided tantalizing clues to support the view that RNA silencing functions as a physiological response to regulate viral infection in vertebrate cells. Amongst these, two manuscripts that are published in recent issues of Science and Immunity, respectively, have provided some of the first direct evidences that RNA silencing is an important component of antiviral defence in vertebrate cells. In addition to demonstrating RNA silencing to be critical to vertebrate innate immunity, these studies also highlight the potential of utilising virus-infection systems as models to refine our understanding on the molecular determinants of RNA silencing in vertebrate cells.
Figures
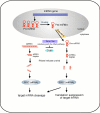
Figure 1
Model of RNA silencing pathway. The biogenesis of RNA silencing transcripts can be derived from either the host cell nucleus mRNA pathway to yield miRNA or the cytoplasmic double strand RNA to yield siRNA. HIV-1 and PFV have evolved to use their transcriptional factor to counteract this ancient host cell immunity.
Similar articles
- Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing.
Bennasser Y, Le SY, Benkirane M, Jeang KT. Bennasser Y, et al. Immunity. 2005 May;22(5):607-19. doi: 10.1016/j.immuni.2005.03.010. Immunity. 2005. PMID: 15894278 - Antiviral innate immune response of RNA interference.
Sidahmed A, Abdalla S, Mahmud S, Wilkie B. Sidahmed A, et al. J Infect Dev Ctries. 2014 Jul 14;8(7):804-10. doi: 10.3855/jidc.4187. J Infect Dev Ctries. 2014. PMID: 25022288 Review. - Induction and suppression of RNA silencing: insights from viral infections.
Voinnet O. Voinnet O. Nat Rev Genet. 2005 Mar;6(3):206-20. doi: 10.1038/nrg1555. Nat Rev Genet. 2005. PMID: 15703763 Review. - Virus counterdefense: diverse strategies for evading the RNA-silencing immunity.
Li F, Ding SW. Li F, et al. Annu Rev Microbiol. 2006;60:503-31. doi: 10.1146/annurev.micro.60.080805.142205. Annu Rev Microbiol. 2006. PMID: 16768647 Free PMC article. Review. - RNA interference-mediated intrinsic antiviral immunity in plants.
Szittya G, Burgyán J. Szittya G, et al. Curr Top Microbiol Immunol. 2013;371:153-81. doi: 10.1007/978-3-642-37765-5_6. Curr Top Microbiol Immunol. 2013. PMID: 23686235 Review.
Cited by
- Dual role of TRBP in HIV replication and RNA interference: viral diversion of a cellular pathway or evasion from antiviral immunity?
Gatignol A, Lainé S, Clerzius G. Gatignol A, et al. Retrovirology. 2005 Oct 27;2:65. doi: 10.1186/1742-4690-2-65. Retrovirology. 2005. PMID: 16253139 Free PMC article. - HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR.
Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. Klase Z, et al. BMC Mol Biol. 2007 Jul 30;8:63. doi: 10.1186/1471-2199-8-63. BMC Mol Biol. 2007. PMID: 17663774 Free PMC article. - HIV-1 Tat interaction with Dicer: requirement for RNA.
Bennasser Y, Jeang KT. Bennasser Y, et al. Retrovirology. 2006 Dec 20;3:95. doi: 10.1186/1742-4690-3-95. Retrovirology. 2006. PMID: 17181864 Free PMC article. - Latency: the hidden HIV-1 challenge.
Marcello A. Marcello A. Retrovirology. 2006 Jan 16;3:7. doi: 10.1186/1742-4690-3-7. Retrovirology. 2006. PMID: 16412247 Free PMC article. Review. - Role of RNA helicases in HIV-1 replication.
Jeang KT, Yedavalli V. Jeang KT, et al. Nucleic Acids Res. 2006;34(15):4198-205. doi: 10.1093/nar/gkl398. Epub 2006 Aug 25. Nucleic Acids Res. 2006. PMID: 16935887 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials