SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli - PubMed (original) (raw)
SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli
Thomas G Bernhardt et al. Mol Cell. 2005.
Abstract
Cell division in Escherichia coli begins with assembly of the tubulin-like FtsZ protein into a ring structure just underneath the cell membrane. Spatial control over Z ring assembly is achieved by two partially redundant negative regulatory systems, the Min system and nucleoid occlusion (NO), which cooperate to position the division site at midcell. In contrast to the well-studied Min system, almost nothing is known about how Z ring assembly is blocked in the vicinity of nucleoids to effect NO. Reasoning that Min function might become essential in cells impaired for NO, we screened for mutations synthetically lethal with a defective Min system (slm mutants). By using this approach, we identified SlmA (Ttk) as the first NO factor in E. coli. Our combined genetic, cytological, and biochemical results suggest that SlmA is a DNA-associated division inhibitor that is directly involved in preventing Z ring assembly on portions of the membrane surrounding the nucleoid.
Figures
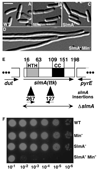
Figure 1. The slmA (ttk) Locus and Phenotypes of slmA Mutants
(A–D) DIC micrographs showing cells of TB43/pTB8 [Δ_laclZYA_::frt_Δ_minCDE::frt/Plao::minCDE::JacZ_] (A and B) and its slmA_127 derivative (C and D). Cells were grown to OD600 = 0.6–0.7 in liquid LB-Amp with (A and C) or without (B and D) 500 µM IPTG and fixed. Bar = 5 µm. (E) Diagram of the slmA locus indicating the location and orientation of the slmA127 and slmA267 EZTnKan-2 insertions (black triangles, inserted after bp 313 or 88 of slmA, respectively). SwissProt annotation P06969 was used to number amino acid residues. Potential DNA binding (HTH) and coiled-coil (CC) regions are indicated. The latter was identified by using COILS (Lupas et al., 1991). The double-headed arrow indicates the endpoints of the Δ_slmA::aph and Δ_slmA::frt deletion-replacement alleles. (F) Overnight cultures of TB28 [wt], TB57 [Para::_minCDE_], TB66 [_slmA127_], and TB68 [slmA127 Para::_minCDE_] were grown to equal density in LB-0.2% arabinose and serially diluted (10−1-10–6) in LB. Aliquots (5 µl) were spotted on LB agar without arabinose and incubated overnight at 30°C.
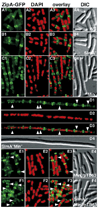
Figure 2. Septal Ring and Nucleoid Positioning in SlmA− and SlmA− Min− Mutants
Shown are representative cells of the following λCH151 [Plac::zipA_-gfp_] lysogens: TB28 [wt] (A), TB85 [Δ_slmA::aph_] (B), TB43 [Δ_minCDE::frt_] (C), TB86 [Δ_slmA::aph Δ_minCDE::_frt_] (D), TB43/pTB63 (E), and TB86/pTB63 (F). Plasmid pTB63 [_ftsQAZ_] is a low-copy, pSC101-derivative expressing ftsQAZ from native promoters. Cells were grown to OD600 = 0.6–0.7 at 30°C in LB with 37 µM IPTG, fixed, stained with DAPI (0.25 µg/ml), and imaged with optics specific for GFP (A1, B1, C1, D1, E1, and F1), DAPI (A2, B2, C2, D2, E2, and F2), and DIC (A4, B4, C4, D4, E4, and F4). A3, B3, C3, D3, E3, and F3 show a digital overlay of the GFP and DAPI signals. Arrowheads point to examples of ZipA-GFP rings, and other types of accumulations, in regions with significant DAPI staining. Bar = 2 µm.
Figure 3. Nucleoid Cutting in SlmA− DnaA− Cells
Cells of TB104 [_cI_857 λPR::dnaA_] (A) and TB105 [Δ_slmA::frt _cI_857 λPR::_dnaA_] (B and C) were grown in LB for 3.5 hr at 30°C to deplete DnaA. Cells were fixed, stained with DAPI, and imaged with DAPI- and DIC-specific optics. A1, B1, and C1 show a digital overlay of the DIC and DAPI images, and A2, B2, and C2 show the DIC image only. Bar = 2 µm. Several parameters of randomly selected cells from each strain were measured, and the results are summarized in (D).
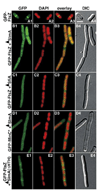
Figure 4. Distribution of GFP-SlmA on the Nucleoid
Shown are live cells of TB85(HKTB99) [Δ_slmA_::frt (Plac::gfp-slmA)] grown to OD600 = 0.5–0.6 at 30°C in LB with 250 µM IPTG. DAPI was added to 0.25 µg/ml 30 min prior to imaging. Cells in (F) and (G) were treated with chloramphenicol (100 µg/ml) and grown for an additional 30 min prior to viewing. Panels show GFP-SlmA (1), DAPI (2), merged (3), and DIC (4) images. Bar = 2 µm.
Figure 5. slmA Overexpression Blocks Z Ring Formation
Shown are cells of strain TB28/pDB407 [wt/λPR::_gfp-ftsZ_] harboring pJF118EH [vector] (A), pTB67 [Ptac::_slmA_] (B), pDR144 [Plac::_sfiA_] (C), or pTB108 [Ptac::slmA(58–198)-_h_] (E). D1–D4 show a cell of strain TB28/pLL14/pTB67 [wt/λPR::gfp-minC(141–231)/Ptac::_slmA_]. Cells were grown to OD600 = 0.25–0.3 at 39°C in LB-Amp-Spc. DAPI (to 0.25 µg/ml) and IPTG (to 50 µM) were added, and growth was continued for 30 min prior to imaging. Cells were imaged live, and panels show GFP (1), DAPI (2), merged (3), and DIC (4) images. Bar = 2 µm. Note that continued incubation of the cells in panels B1–E4 led to the formation of very long nonseptate filaments, indicative of a complete division block.
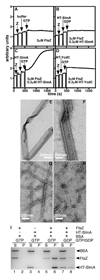
Figure 6. Formation of FtsZ-SlmA Ribbons
(A–D) 90° angle light scattering was monitored in reactions containing 3 µM FtsZ before and after the addition of HT-SlmA storage buffer and GTP (1 mM) (A), HT-SlmA (0.3 µM) and GDP (1 mM) (B), HT-SlmA (0.3 µM) and GTP (1 mM) (C), and HT-YcdC (0.3 µM) and GTP (1 mM) (D). (E–H) Electron micrographs of structures formed in reactions containing 3 µM FtsZ and 1 mM GTP with the addition of 0.6 µM HT-SlmA (E and F), 0.6 µM HT-YcdC (G), or HT-SlmA storage buffer (H). Bar = 500 nm (E) or 200 nm (F–H). (I)SDS-PAGE analysis of supernatant (S) and pellet (P) fractions after centrifugation of reactions containing nucleotide (1 mM) and proteins (6 µM each) as indicated. Note that the pellet fractions were twice as concentrated as the supernatant fractions (see Experimental Procedures).
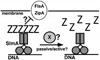
Figure 7. Model for SlmA Function
DNA bound SlmA, presumed to be a dimer, competes with membrane bound septal ring components, such as ZipA and FtsA, for binding FtsZ polymers. In subsequent steps, SlmA may play a rather passive role in that SlmA bound polymers may simply turn-over due to their intrinsic GTP hydrolytic activity. A more attractive possibility is that SlmA, perhaps in combination with some other factor (X), actively promotes the disassembly of the FtsZ polymers.
Similar articles
- The Nucleoid Occlusion Protein SlmA Binds to Lipid Membranes.
Robles-Ramos MÁ, Margolin W, Sobrinos-Sanguino M, Alfonso C, Rivas G, Monterroso B, Zorrilla S. Robles-Ramos MÁ, et al. mBio. 2020 Sep 1;11(5):e02094-20. doi: 10.1128/mBio.02094-20. mBio. 2020. PMID: 32873767 Free PMC article. - SlmA antagonism of FtsZ assembly employs a two-pronged mechanism like MinCD.
Du S, Lutkenhaus J. Du S, et al. PLoS Genet. 2014 Jul 31;10(7):e1004460. doi: 10.1371/journal.pgen.1004460. eCollection 2014 Jul. PLoS Genet. 2014. PMID: 25078077 Free PMC article. - Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli.
Bailey MW, Bisicchia P, Warren BT, Sherratt DJ, Männik J. Bailey MW, et al. PLoS Genet. 2014 Aug 7;10(8):e1004504. doi: 10.1371/journal.pgen.1004504. eCollection 2014 Aug. PLoS Genet. 2014. PMID: 25101671 Free PMC article. - Bacterial Nucleoid Occlusion: Multiple Mechanisms for Preventing Chromosome Bisection During Cell Division.
Schumacher MA. Schumacher MA. Subcell Biochem. 2017;84:267-298. doi: 10.1007/978-3-319-53047-5_9. Subcell Biochem. 2017. PMID: 28500529 Review. - Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring.
Lutkenhaus J. Lutkenhaus J. Annu Rev Biochem. 2007;76:539-62. doi: 10.1146/annurev.biochem.75.103004.142652. Annu Rev Biochem. 2007. PMID: 17328675 Review.
Cited by
- A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli.
Espéli O, Borne R, Dupaigne P, Thiel A, Gigant E, Mercier R, Boccard F. Espéli O, et al. EMBO J. 2012 May 11;31(14):3198-211. doi: 10.1038/emboj.2012.128. EMBO J. 2012. PMID: 22580828 Free PMC article. - The Min system and other nucleoid-independent regulators of Z ring positioning.
Rowlett VW, Margolin W. Rowlett VW, et al. Front Microbiol. 2015 May 13;6:478. doi: 10.3389/fmicb.2015.00478. eCollection 2015. Front Microbiol. 2015. PMID: 26029202 Free PMC article. Review. - FtsZ Interactions and Biomolecular Condensates as Potential Targets for New Antibiotics.
Zorrilla S, Monterroso B, Robles-Ramos MÁ, Margolin W, Rivas G. Zorrilla S, et al. Antibiotics (Basel). 2021 Mar 4;10(3):254. doi: 10.3390/antibiotics10030254. Antibiotics (Basel). 2021. PMID: 33806332 Free PMC article. Review. - Cell division is antagonized by the activity of peptidoglycan endopeptidases that promote cell elongation.
Truong TT, Vettiger A, Bernhardt TG. Truong TT, et al. Mol Microbiol. 2020 Dec;114(6):966-978. doi: 10.1111/mmi.14587. Epub 2020 Aug 31. Mol Microbiol. 2020. PMID: 32866331 Free PMC article. - Addiction toxin Fst has unique effects on chromosome segregation and cell division in Enterococcus faecalis and Bacillus subtilis.
Patel S, Weaver KE. Patel S, et al. J Bacteriol. 2006 Aug;188(15):5374-84. doi: 10.1128/JB.00513-06. J Bacteriol. 2006. PMID: 16855226 Free PMC article.
References
- Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 2005;55:1631–1645. - PubMed
- Aussel L, Barre F-X, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases