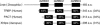Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells - PubMed (original) (raw)
Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells
Kuniaki Saito et al. PLoS Biol. 2005 Jul.
Abstract
microRNAs (miRNAs) are a large family of 21- to 22-nucleotide non-coding RNAs that interact with target mRNAs at specific sites to induce cleavage of the message or inhibit translation. miRNAs are excised in a stepwise process from primary miRNA (pri-miRNA) transcripts. The Drosha-Pasha/DGCR8 complex in the nucleus cleaves pri-miRNAs to release hairpin-shaped precursor miRNAs (pre-miRNAs). These pre-miRNAs are then exported to the cytoplasm and further processed by Dicer to mature miRNAs. Here we show that Drosophila Dicer-1 interacts with Loquacious, a double-stranded RNA-binding domain protein. Depletion of Loquacious results in pre-miRNA accumulation in Drosophila S2 cells, as is the case for depletion of Dicer-1. Immuno-affinity purification experiments revealed that along with Dicer-1, Loquacious resides in a functional pre-miRNA processing complex, and stimulates and directs the specific pre-miRNA processing activity. These results support a model in which Loquacious mediates miRNA biogenesis and, thereby, the expression of genes regulated by miRNAs.
Figures
Figure 1. Loqs/CG6866 Is a Paralog of R2D2
A protein sequence alignment of Loqs, R2D2, and RDE-4 (C. elegans). The two canonical dsRBDs are boxed. Conserved residues are shaded in gray. It is noted that the C-terminal region of Loqs contains a non-canonical dsRBD (see Figure 8).
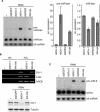
Figure 2. Depletion of Loqs and Dicer-1 Causes Pre-miRNA Accumulation
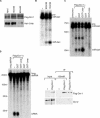
(A) dsRNAs of Loqs, Dicer-1 (Dcr-1), Dicer-2 (Dcr-2), R2D2, or EGFP were introduced to S2 cells by soaking. Total RNA was isolated 4 d after initial exposure of S2 cells to the indicated target genes. The RNA was separated on a 12% denaturing polyacrylamide gel, transferred to a nylon membrane, and probed with 5′-radiolabeled miR-bantam (miR-ban) antisense oligodeoxynucleotide (top panel) and re-probed for U6 snRNA (bottom panel). After normalization for loading, the relative ratios for pre-miRNA (pre-miR-ban) (left panel) and mature miRNA (miR-ban) (right panel) were calculated and normalized to EGFP dsRNA experiments. Depletion of Dicer-1 (Dcr-1) by RNAi resulted in a marked accumulation of pre-miR-ban and a modest reduction in levels of mature miR-ban. (B) Upper panel: Analysis of transcript levels using RT-PCR for Loqs and Dicer-1 (Dcr-1) after treatment of Drosophila S2 cells with dsRNAs against each protein. AGO2 was used as control. Lower panel: Loqs dsRNA did not affect Dicer-1 protein levels. dsRNAs of Loqs, Dicer-1, or EGFP were introduced to S2 cells by soaking. After 4 d, cell lysates were prepared and the levels of Dicer-1 protein were measured by Western blotting. (C) As in (A), total RNA was probed with 5′-radiolabeled miR-8 antisense oligodeoxynucleotide (top panel) and re-probed for U6 snRNA (bottom panel).
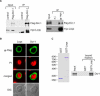
Figure 3. Loqs Associates with Dicer-1
(A) Loqs and Dicer-1 form a complex in vivo. Left panel: Protein extract was prepared from S2 cells that expressed both full-length myc-tagged Loqs (myc-Loqs) and Flag-tagged Dicer-1 (Flag-Dcr-1). Protein extract containing only myc-Loqs was also prepared (control). Total extracts (input) and the materials obtained after immunoprecipitation from the extracts with anti-Flag were run on an SDS-polyacrylamide gel. Western blots were prepared and immunostained with anti-myc (to detect myc-Loqs) or anti-Flag (to detect Flag–Dicer-1) antibodies. The protein band shown by an asterisk is an antibody used. Right panel: Protein extracts were prepared from S2 cells that expressed both myc-Loqs and Flag–Dicer-1, and immunoprecipitation was performed with anti-myc antibody (α-myc). Non-specific antisera were also employed as a negative control (n.i.). Flag–Dicer-1 was specifically co-immunoprecipitated with myc-Loqs. (B) Immunofluorescence using anti-Flag antibodies show that both Loqs and Dicer-1 are predominantly localized in the cytoplasm in Drosophila S2 cells (α-Flag). Flag–Dicer-1 (Dcr-1) and Flag–Loqs were transiently expressed in the cells by transfection. The nuclear DNA was stained with propidium iodide. A DIC image of the same field is also shown (DIC). (C) Loqs interacts with Dicer-1 in an RNase-resistant manner in vitro. 35S-labeled Dicer-1 was produced by an in vitro transcription and translation system in the presence of [35S]methionine, treated with RNaseA, and incubated with either GST–Loqs or GST itself immobilized on glutathione-Sepharose resins. After extensive washing, the bound fractions were resolved on an SDS-polyacrylamide gel and the protein labeled with 35S visualized by autoradiography. The Coomassie Blue stainings of GST and GST–Loqs used in this experiment are shown on the left.
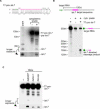
Figure 4. Synthetic D. melanogaster let-7 Precursor Processing in the Cytoplasmic Lysate of S2 Cells
(A) S2 cytoplasmic lysate is capable of processing pre-let-7 into mature let-7. Pre-let-7 (the sequence shown on the top) was transcribed with T7 RNA polymerase in vitro (T7-pre-let-7). After gel purification, it was incubated with S2 cytoplasmic lysate, and resultant RNA was subjected to Northern blotting using an oligodeoxynucleotide recognizing both pre-let-7 and mature let-7. Total RNA prepared from pupa was also applied (pupa), which shows where endogenous pre-let-7 (pre-let-7; ∼70 nucleotides) and mature let-7 (let-7; 21 nucleotides) migrate on the blot. The cytoplasmic lysate (+) lane, but not (−) lane, shows a band corresponding to mature_let-7,_ meaning that the synthetic T7-pre-let-7 was processed to the matured form in the lysate. (B) Functional analysis of mature let-7 produced from synthetic T7-pre-let-7 in (A). Cap-labeled target RNA with let-7 target site (∼500 nucleotides) was incubated in S2 cytoplasmic lysate with or without T7-pre-let-7 for 3 h._let-7_-directed cleavage product (∼150 mucleotides) was observed only when T7-pre-let-7 was included, indicating that let-7 produced in the lysate is functional. (C) Dicer-1- or Loqs-depleted cytoplasmic lysate has less activity to produce mature let-7 from the precursor. Dicer-1 (Dcr-1) or Loqs was depleted from S2 cells by RNAi, and the cytoplasmic lysate was assayed for pre-let-7 processing as in (A). "-" indicates no lysate.
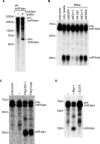
Figure 5. In Vitro Processing Activities of Loqs and Dicer-1
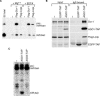
(A) Preparation of pre-miR-ban. Uniformly labeled pri-miR-ban was incubated with S2 nuclear lysate for the processing. The resultant pre-miR-ban fragment (pre-miR-ban) was gel-purified and used as a substrate in the pre-miRNA processing assays. (B) In vitro processing of pre-miR-ban using S2 cytoplasmic lysates. Cytoplasmic lysates were prepared 4 d after the initial exposure of S2 cells to the indicated target genes (as in Figure 2A) and used for in vitro processing. The gel-purified pre-miR-ban was incubated in cytoplasmic lysates for 1 h. “cyto. lysate” indicates parental S2 cytoplasmic lysate that shows activity for generating mature miR-ban from the gel-purified pre-miR-ban in (A). (C) In vitro processing of pre-miR-ban using immunopurified Dicer-1 and Loqs complexes. Purified Flag–Dicer-1 (Flag-Dcr-1) and Flag–Loqs complexes were incubated with pre-miR-ban for 2 h and tested for processing activity. “−” shows the activity of a negative control prepared from parental S2 cells. (D) The pre-miRNA processing activity of Flag–Dicer-1 complex in the presence and absence of magnesium ions. Purified Flag–Dicer-1 complex was incubated with pre-miR-ban with or without magnesium ions in buffers. Addition of EDTA caused the abolition of the activity.
Figure 6. Loqs Stimulates the Specific Processing of Pre-miRNA by Dicer-1
(A) Flag–Dicer-1 complex was purified under a harsh condition. Protein extract was prepared from S2 cells that expressed both full-length myc-tagged Loqs (myc-Loqs) and Flag-tagged Dicer-1 (Flag-Dcr-1). The amounts of Loqs in Flag–Dicer-1 complexes prepared under high-salt condition and low-salt condition were examined by Western blotting using anti-myc antibody, which show that less Loqs was co-purified with Dicer-1 in the high-salt condition. (B) The miRNA processing activities of Flag–Dicer-1 (Flag-Dcr-1) complexes in (A). Flag–Dicer-1 complex containing less Loqs showed a lower activity for the processing. (C) Recombinant Loqs stimulates the in vitro processing of pre-miR-ban by Flag–Dicer-1 complex purified in high-salt condition. 100 ng of purified GST or GST–Loqs (see Figure 3C) were supplemented for the processing activity by Flag–Dicer-1 complex. GST–Loqs by itself does not show any pre-miRNA processing activity. (D) GST–Loqs inhibits the siRNA-generating activity of Dicer-1. Uniformly labeled long dsRNA was incubated with Flag–Dicer-1 (Flag-Dcr-1) complex purified in high-salt condition in (A) with or without GST–Loqs. The Flag–Dicer-1 complex by itself showed a considerable activity of generating siRNA from long dsRNA. Addition of GST–Loqs, but not GST, inhibited the processing. Note that the Flag–Dicer-1 (Flag-Dcr-1) complex does not contain Dicer-2 (Dcr-2), judged by Western blot analysis using anti-Dicer-2 antibodies (right panel).
Figure 7. Dicer-1–Loqs Complexes Are Associated with Pre-miRNA and Mature miRNA In Vivo
(A) Northern blot analyses show that Flag–Dicer-1 (Flag-Dcr-1) complex contains both pre- and mature form of miR-ban. Notably, the precursor form of miR-ban was accumulated in the Dicer-1 complex. In the case of the Loqs complex, the precursor apparently accumulated within the complex when the Flag–Loqs complex was prepared in the presence of EDTA that was shown to inhibit the pre-miRNA processing activity in Figure 5D. (B) AGO1 associates with Dicer-1 and Loqs. IgG-bound fractions prepared from S2 cells expressing AGO1–TAP, EGFP–TAP, or the parental S2 cells (−), were subjected to Western blotting using antibodies against Dicer-1, AGO1, and Flag (for Loqs). Dicer-1 is not present in AGO2-associated complex. (C) In vitro processing of pre-miR-ban using affinity-purified AGO1 complexes. Purified AGO1–TAP or AGO2–TAP complexes were incubated with pre-miR-ban (precursor) and tested for processing activity. “−” shows the activity of a negative control prepared from parental S2 cells.
Figure 8. Domain Structure of Loqs and Its_Homo sapiens_ and Xenopus Homologs
The dsRNA-binding motif (dsRBD) is indicated as a black box. These proteins contain three putative dsRBDs. Loqs shares ∼34% amino acid identity with TRBP and PACT and ∼31% identity with Xlrbpa (Xenopus laevis RNA-binding protein A). Sequence comparison between Loqs and its human and Xenopus homologs also showed a higher degree of amino acid conservation in dsRBDs including C-terminal non-canonical dsRBDs. The_Xenopus_ homolog, Xlrbpa, of TRBP/PACT has been found to associate with ribosomes in the cytoplasm [77], as is the case for many RNAi factors including miRNAs [47,78−82].
Similar articles
- Genome-wide identification of targets of the drosha-pasha/DGCR8 complex.
Kadener S, Rodriguez J, Abruzzi KC, Khodor YL, Sugino K, Marr MT 2nd, Nelson S, Rosbash M. Kadener S, et al. RNA. 2009 Apr;15(4):537-45. doi: 10.1261/rna.1319309. Epub 2009 Feb 17. RNA. 2009. PMID: 19223442 Free PMC article. - Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein.
Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Förstemann K, et al. PLoS Biol. 2005 Jul;3(7):e236. doi: 10.1371/journal.pbio.0030236. Epub 2005 May 24. PLoS Biol. 2005. PMID: 15918770 Free PMC article. - The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis.
Landthaler M, Yalcin A, Tuschl T. Landthaler M, et al. Curr Biol. 2004 Dec 14;14(23):2162-7. doi: 10.1016/j.cub.2004.11.001. Curr Biol. 2004. PMID: 15589161 - MicroRNA biogenesis: isolation and characterization of the microprocessor complex.
Gregory RI, Chendrimada TP, Shiekhattar R. Gregory RI, et al. Methods Mol Biol. 2006;342:33-47. doi: 10.1385/1-59745-123-1:33. Methods Mol Biol. 2006. PMID: 16957365 Review. - Orientation of Human Microprocessor on Primary MicroRNAs.
Nguyen HM, Nguyen TD, Nguyen TL, Nguyen TA. Nguyen HM, et al. Biochemistry. 2019 Jan 29;58(4):189-198. doi: 10.1021/acs.biochem.8b00944. Epub 2018 Dec 10. Biochemistry. 2019. PMID: 30481000 Review.
Cited by
- MicroRNA-Mediated Host-Pathogen Interactions Between Bombyx mori and Viruses.
Awais MM, Shakeel M, Sun J. Awais MM, et al. Front Physiol. 2021 May 7;12:672205. doi: 10.3389/fphys.2021.672205. eCollection 2021. Front Physiol. 2021. PMID: 34025458 Free PMC article. Review. - A Glimpse of "Dicer Biology" Through the Structural and Functional Perspective.
Paturi S, Deshmukh MV. Paturi S, et al. Front Mol Biosci. 2021 May 7;8:643657. doi: 10.3389/fmolb.2021.643657. eCollection 2021. Front Mol Biosci. 2021. PMID: 34026825 Free PMC article. Review. - Signature miRNAs involved in the innate immunity of invertebrates.
Yang G, Yang L, Zhao Z, Wang J, Zhang X. Yang G, et al. PLoS One. 2012;7(6):e39015. doi: 10.1371/journal.pone.0039015. Epub 2012 Jun 19. PLoS One. 2012. PMID: 22723921 Free PMC article. - Regulation of microRNA biogenesis and turnover by animals and their viruses.
Libri V, Miesen P, van Rij RP, Buck AH. Libri V, et al. Cell Mol Life Sci. 2013 Oct;70(19):3525-44. doi: 10.1007/s00018-012-1257-1. Epub 2013 Jan 26. Cell Mol Life Sci. 2013. PMID: 23354060 Free PMC article. Review. - microRNA strand selection: Unwinding the rules.
Medley JC, Panzade G, Zinovyeva AY. Medley JC, et al. Wiley Interdiscip Rev RNA. 2021 May;12(3):e1627. doi: 10.1002/wrna.1627. Epub 2020 Sep 20. Wiley Interdiscip Rev RNA. 2021. PMID: 32954644 Free PMC article. Review.
References
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403:901–906. - PubMed
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. - PubMed
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila . Cell. 2003;113:25–36. - PubMed
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases