A new approach for the analysis of bacterial microarray-based Comparative Genomic Hybridization: insights from an empirical study - PubMed (original) (raw)
A new approach for the analysis of bacterial microarray-based Comparative Genomic Hybridization: insights from an empirical study
Eduardo N Taboada et al. BMC Genomics. 2005.
Abstract
Background: Microarray-based Comparative Genomic Hybridization (M-CGH) has been used to characterize the extensive intraspecies genetic diversity found in bacteria at the whole-genome level. Although conventional microarray analytical procedures have proved adequate in handling M-CGH data, data interpretation using these methods is based on a continuous character model in which gene divergence and gene absence form a spectrum of decreasing gene conservation levels. However, whereas gene divergence may yet be accompanied by retention in gene function, gene absence invariably leads to loss of function. This distinction, if ignored, leads to a loss in the information to be gained from M-CGH data. We present here results from experiments in which two genome-sequenced strains of C. jejuni were compared against each other using M-CGH. Because the gene content of both strains was known a priori, we were able to closely examine the effects of sequence divergence and gene absence on M-CGH data in order to define analytical parameters for M-CGH data interpretation. This would facilitate the examination of the relative effects of sequence divergence or gene absence in comparative genomics analyses of multiple strains of any species for which genome sequence data and a DNA microarray are available.
Results: As a first step towards improving the analysis of M-CGH data, we estimated the degree of experimental error in a series of experiments in which identical samples were compared against each other by M-CGH. This variance estimate was used to validate a Log Ratio-based methodology for identification of outliers in M-CGH data. We compared two genome strains by M-CGH to examine the effect of probe/target identity on the Log Ratios of signal intensities using prior knowledge of gene divergence and gene absence to establish Log Ratio thresholds for the identification of absent and conserved genes.
Conclusion: The results from this empirical study validate the Log Ratio thresholds that have been used in other studies to establish gene divergence/absence. Moreover, the analytical framework presented here enhances the information content derived from M-CGH data by shifting the focus from divergent/absent gene detection to accurate detection of conserved and absent genes. This approach closely aligns the technical limitations of M-CGH analysis with practical limitations on the biological interpretation of comparative genomics data.
Figures
Figure 1
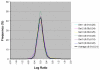
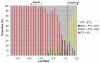
Log Ratio distribution of self-self experiments. The LR distribution of self-self experiments was used to determine the level of experimental variability in our experimental platform. Standardized samples of genomic DNA from C. jejuni NCTC 11168 (or RM1221) labelled with Cy3 and Cy5 were co-hybridized to our microarray. Results from six replicates had mean LR = 0.01 with an average SD of 0.215. Because samples from the same genomic DNA were used in both channels, LRs were expected to remain close to 0 and any deviations could be attributable to experimental error.
Figure 2
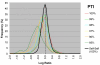
Log Ratio distributions of highly conserved genes. LR distributions from a series of M-CGH experiments comparing two genome-sequenced strains of C. jejuni (NCTC 11168 vs. RM1221). Genes were binned according to PTI and the LR distributions of bins with greater than 95% PTI are presented here. Because the microarray was designed based on strain NCTC 11168, LR deviations from 0 would be the result of sequence divergence or gene absence in strain RM1221. The LR distributions of genes with greater than 99% PTI do not deviate significantly from the average distribution of a Self-self experiment whereas increasingly larger deviations are observed in the range from 98 to 95% PTI.
Figure 3
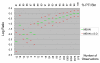
Relationship between Log Ratio and PTI. We plotted the average LR of genes with varying levels of % PTI from M-CGH experiments comparing two genome-sequenced strains of C. jejuni (NCTC 11168 vs. RM1221). Genes were binned according to PTI and the LR distributions of individual bins are presented here. The number of observations made within each PTI bin is shown in the lower axis. As seen in Figure 2, the average LR of genes with high levels of PTI are very similar, although a noticeable decrease in average LR is observed even at 98% PTI. Although the average LR becomes increasingly negative as PTI levels drop, given the SD observed within each group PTI category, there is considerable overlap between categories.
Figure 4
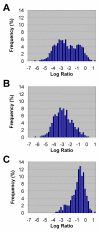
Log Ratio distribution of genes absent from strain RM1221. We plotted the LR distribution of genes predicted to be absent from strain RM1221 based on BLAST searches of NCTC 11168 against the RM1221 genome. Because of the lack of Tester signal predicted from these genes, LRs should be expected to be highly negative. The LR distribution (A) appeared to be bimodal with a significant number of genes bearing unusually high LRs. Further examination of these genes revealed a potential bias towards genes represented by short probes on the microarray (i.e. less than 250 bp). Separate re-plotting of the LR distributions of long (B) and short (C) probes confirms a higher average LR among genes with short probes.
Figure 5
Determination of thresholds for absent and conserved genes. We calculated the proportion of genes belonging to each of four PTI categories at 0.2 LR intervals in order to determine LR thresholds that could be used to predict absent and conserved genes with a high degree of certainty. Below a LR of -3.0, the false positive rate for conserved genes is less than 1%; similarly, the false positive rate for absent genes above LRs of -0.8 is also less than 1%. In the LR interval between -3.0 and -0.8, particularly approaching the -0.8 boundary, there are significant number of genes from more than one PTI category and thus there is significant risk of misclassification.
Similar articles
- A process for analysis of microarray comparative genomics hybridisation studies for bacterial genomes.
Carter B, Wu G, Woodward MJ, Anjum MF. Carter B, et al. BMC Genomics. 2008 Jan 29;9:53. doi: 10.1186/1471-2164-9-53. BMC Genomics. 2008. PMID: 18230148 Free PMC article. - Large-scale comparative genomics meta-analysis of Campylobacter jejuni isolates reveals low level of genome plasticity.
Taboada EN, Acedillo RR, Carrillo CD, Findlay WA, Medeiros DT, Mykytczuk OL, Roberts MJ, Valencia CA, Farber JM, Nash JH. Taboada EN, et al. J Clin Microbiol. 2004 Oct;42(10):4566-76. doi: 10.1128/JCM.42.10.4566-4576.2004. J Clin Microbiol. 2004. PMID: 15472310 Free PMC article. - Characterization of a novel Lactobacillus species closely related to Lactobacillus johnsonii using a combination of molecular and comparative genomics methods.
Sarmiento-Rubiano LA, Berger B, Moine D, Zúñiga M, Pérez-Martínez G, Yebra MJ. Sarmiento-Rubiano LA, et al. BMC Genomics. 2010 Sep 17;11:504. doi: 10.1186/1471-2164-11-504. BMC Genomics. 2010. PMID: 20849602 Free PMC article. - Microarray-based comparative genomic hybridization and its applications in human genetics.
Oostlander AE, Meijer GA, Ylstra B. Oostlander AE, et al. Clin Genet. 2004 Dec;66(6):488-95. doi: 10.1111/j.1399-0004.2004.00322.x. Clin Genet. 2004. PMID: 15521975 Review. - Recent advances in array comparative genomic hybridization technologies and their applications in human genetics.
Lockwood WW, Chari R, Chi B, Lam WL. Lockwood WW, et al. Eur J Hum Genet. 2006 Feb;14(2):139-48. doi: 10.1038/sj.ejhg.5201531. Eur J Hum Genet. 2006. PMID: 16288307 Review.
Cited by
- A process for analysis of microarray comparative genomics hybridisation studies for bacterial genomes.
Carter B, Wu G, Woodward MJ, Anjum MF. Carter B, et al. BMC Genomics. 2008 Jan 29;9:53. doi: 10.1186/1471-2164-9-53. BMC Genomics. 2008. PMID: 18230148 Free PMC article. - Epidemiology, relative invasive ability, molecular characterization, and competitive performance of Campylobacter jejuni strains in the chicken gut.
Pope C, Wilson J, Taboada EN, Mackinnon J, Felipe Alves CA, Nash JH, Rahn K, Tannock GW. Pope C, et al. Appl Environ Microbiol. 2007 Dec;73(24):7959-66. doi: 10.1128/AEM.01657-07. Epub 2007 Oct 5. Appl Environ Microbiol. 2007. PMID: 17921281 Free PMC article. - Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization.
LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. LoCascio RG, et al. Appl Environ Microbiol. 2010 Nov;76(22):7373-81. doi: 10.1128/AEM.00675-10. Epub 2010 Aug 27. Appl Environ Microbiol. 2010. PMID: 20802066 Free PMC article. - Microarray-based comparative genomic profiling of reference strains and selected Canadian field isolates of Actinobacillus pleuropneumoniae.
Gouré J, Findlay WA, Deslandes V, Bouevitch A, Foote SJ, MacInnes JI, Coulton JW, Nash JH, Jacques M. Gouré J, et al. BMC Genomics. 2009 Feb 24;10:88. doi: 10.1186/1471-2164-10-88. BMC Genomics. 2009. PMID: 19239696 Free PMC article. - Comparative and functional genomics reveals genetic diversity and determinants of host specificity among reference strains and a large collection of Chinese isolates of the phytopathogen Xanthomonas campestris pv. campestris.
He YQ, Zhang L, Jiang BL, Zhang ZC, Xu RQ, Tang DJ, Qin J, Jiang W, Zhang X, Liao J, Cao JR, Zhang SS, Wei ML, Liang XX, Lu GT, Feng JX, Chen B, Cheng J, Tang JL. He YQ, et al. Genome Biol. 2007;8(10):R218. doi: 10.1186/gb-2007-8-10-r218. Genome Biol. 2007. PMID: 17927820 Free PMC article.
References
- Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–80. doi: 10.1038/16495. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials