PIASx acts as an Elk-1 coactivator by facilitating derepression - PubMed (original) (raw)
PIASx acts as an Elk-1 coactivator by facilitating derepression
Shen-Hsi Yang et al. EMBO J. 2005.
Abstract
The ETS-domain transcription factor Elk-1 is a MAP kinase-inducible transcriptional activator protein. However, in the basal state, its activity is repressed by SUMO-dependent histone deacetylase (HDAC) recruitment. Relief of this repression accompanies the activation process. Here, we demonstrate that PIASx(alpha) acts to facilitate this derepression process. Members of the PIAS family of proteins can act as E3 enzymes that enhance the sumoylation status of a variety of substrates. However, PIASx-mediated coactivation of Elk-1 occurs in an E3 activity-independent manner. PIASx(alpha) binds to Elk-1 in vivo and enhances its transcriptional activity. The coactivating properties of PIASx(alpha) require Elk-1 to be modified with SUMO and the integrity of the SUMO binding motif in PIASx(alpha). PIASx(alpha) activates Elk-1 through alterations in the HAT/HDAC activities associated with Elk-1. In particular, PIASx(alpha) facilitates the loss of the repressive HDAC-2 from sumoylated Elk-1, a key event in the activation of Elk-1 in response to signalling through the ERK MAP kinase pathway. Our data therefore reveal a novel coactivator function for PIASx(alpha) through reversing SUMO-mediated repression of transcription factor activity.
Figures
Figure 1
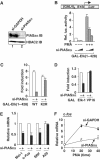
siRNAs show that endogenous PIASxα is a coactivator of Elk-1. (A) Western blot showing a reduction in PIASxα levels in the presence of specific RNAi duplexes. Total lysates from HeLa cells transfected with siRNAs against either GAPDH or _PIASx_α were probed with PIASxα or HDAC-2 antibodies. (B–D) Reporter gene analyses of the activities of the indicated GAL fusion proteins in 293 cells in the presence of GAPDH (−) or _PIASx_α RNAi duplexes. (B) The activity of GAL-Elk(1–428) in the presence or absence of PMA stimulation as indicated and siRNAs against either GAPDH (−) (100 pmol) or increasing amounts of _PIASx_α (50 and 100 pmol). (C) Activities of the wild-type (WT) and mutant (K2R) GAL-Elk(1–428) in the presence of PMA and cells in the presence of GAPDH (−) or _PIASx_α RNAi duplexes. Data are shown as relative luciferase activity relative to GAL-Elk(1–428) plus GAPDH (−) RNAi duplexes, in the absence of PMA, taken as 1 (B, C). (D) Activity of GAL-Elk(1–428) and GAL-VP16 fusion proteins in the presence of GAPDH (−) or _PIASx_α RNAi duplexes. Data are shown relative to the activity of each construct in the absence of RNAi duplexes against PIASxα. Luciferase assays are representative of at least two independent experiments (standard errors are shown; _n_=2). (E, F) Real-time RT–PCR analysis of the indicated genes in serum-starved (E) or PMA-stimulated (F) HeLa cells in the presence or absence of RNAi duplexes to either GAPDH (−) or PIASxα. PMA stimulation was for the indicated times. Data are averages of two experiments performed in duplicate.
Figure 2
Activation of sumoylated Elk-1 by PIASxα. Reporter gene analysis of the activities of GAL-Elk fusion proteins on the GAL-driven E1b promoter-reporter construct (schematic shown in [A]) in 293 cells is shown. (A) The activity of GAL-Elk(1–428) was tested in the presence and absence of cotransfected PIASxα (0.1 μg), PIASxβ (0.1 μg), PIAS1 (0, 0.1, 0.5 and 1 μg) and PIASy (0, 0.1, 0.5 and 1 μg) as indicated and presented as fold induction of reporter activity relative to the absence of PIAS proteins. (B) The activities of the wild-type (WT) and mutant (K2R) GAL-Elk(1–428) fusion proteins were tested in the presence or absence of increasing amounts of PIASxα (0, 10, 25 and 100 ng) as indicated and presented as fold induction of reporter activity relative to the absence of PIASxα (taken as 1). For alternative depiction of data, see Supplementary Figure S1. (C) The activities of GAL-Elk(1–428) WT and K2R were tested in the presence or absence of PIASxα (0.25 μg) and/or DN-ubc9 (1 μg) as indicated. The data are presented as fold induction by PIASxα of reporter activity and are calculated from the activity of the reporter under each condition relative to the activity in the absence of PIASxα. (D) The activities of GAL-Elk(260–428) and GAL-Sumo-Elk(260–428) were tested in the presence or absence of PIASxα (0.25 μg). The data are presented as fold induction by PIASxα of reporter activity and are relative to the activity of each construct in the absence of PIASxα. (E, F) Activities of GAL-Elk(223–428), GAL-SAP-1(234–431) (E), GAL-DBD, GAL-Elk(1–428) and GAL-VP16 (F) in 293 cells in the presence or absence of transfected PIASxα. Data are shown as fold induction relative to the activity of each GAL fusion in the absence of PIASxα. Luciferase assays are representative of at least two independent experiments (standard errors are shown; _n_=2).
Figure 3
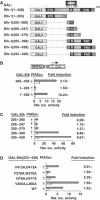
Mapping the determinants in Elk-1 for PIASxα coactivation. (A) Schematic of the GAL fusion proteins used in this figure. The Elk-1 ETS domain, R-motif and TAD are indicated in dark grey boxes. (B–D) Reporter gene analysis of activities of indicated GAL-Elk fusion proteins on the GAL-driven E1b promoter-reporter construct in 293 cells. The activities of each construct were tested in the presence or absence of transfected PIASxα and presented as relative luciferase activities (relative to activities of GAL-Elk(1–428) (B), GAL-Elk(205–428) (C) and GAL-Elk(223–428) (D) in the absence of PIASxα) and _n_-fold induction by PIASxα. Luciferase assays are representative of at least two independent experiments (standard errors are shown; _n_=2).
Figure 4
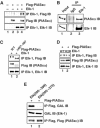
Elk-1 and PIASxα interact in vivo. (A) Co-immunoprecipitation (IP) of the indicated PIASxα proteins with Elk-1 from 293 cells. Cells were cotransfected with 2 μg of wild-type (WT) or mutant (K2R) pcDNA-Elk-1 and 3 μg of WT or mutant (Δ467–487) pFlag-PIASxα as indicated. Elk-1 proteins were immunoprecipitated with an anti-Elk-1 antibody, and co-precipitated PIASxα proteins were subsequently detected with a Flag antibody (top panel). (B) Co-immunoprecipitation of endogenous Elk-1 and PIASxα from HeLa cells. Immunoprecipitation (IP) was carried out using Elk-1 or control IgG antibodies. (C, D) Co-immunoprecipitation (IP) of the indicated PIASxα proteins with Elk-1 was carried out as in (A), except that 2 μg of mutant (K2R) pcDNA-Elk-1 and 3 μg of mutant (Δ467–487) pFlag-PIASxα were also tested as indicated. (E) Co-immunoprecipitation (IP) of the indicated GAL-Elk fusion proteins with PIASxα from 293 cells. Cells were cotransfected with 2 μg of each GAL-Elk fusion protein and 3 μg of pFlag-PIASxα. PIASxα was immunoprecipitated with an anti-Flag antibody, and co-precipitated GAL-Elk fusion proteins were subsequently detected with a GAL antibody (top panel). Immunoprecipitates (bottom panel) and total cell extracts (middle panel) were also analysed by immunoblot with the indicated antibodies to detect total levels of proteins in the immunoprecipitate and total cell lysates, respectively.
Figure 5
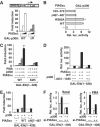
PIASxα effects on Elk-1 sumoylation and phosphorylation status. (A) In vivo sumoylation of Elk-1 in COS7 cells. The indicated His-tagged Elk-1- and HA-tagged SUMO-1-encoding constructs were cotransfected in the presence or absence of increasing amounts of PIASxα (0.25, 1 and 2 μg) or DN-ubc9 (0.25, 1 and 2 μg) plasmids. Elk-1 was isolated by nickel affinity chromatography, and SUMO-modified Elk-1 was detected with anti-HA antibody. Total Elk-1 was detected by anti-Flag antibody. (B) Western blots of extracts from cells transfected with Elk-1 and/or PIASxα in the presence or absence of PMA, probed with an anti-phospho-Ser383 Elk-1 antibody (top panel) and an anti-Flag antibody (bottom panel). (C) Reporter gene analysis of the activity of GAL-Elk(1–428) in the presence or absence of PIASxα and/or PMA and/or MEK inhibitor (U01260) on the GAL-driven E1B promoter-reporter construct in 293 cells. The data are shown relative to GAL-Elk(1–428) in the absence of PMA and PIASxα. (D) In vivo sumoylation of Elk-1 in HeLa cells. His-tagged Elk-1 and HA-tagged SUMO-1 were cotransfected in the absence and presence of PIASxα siRNAs. Cells were either serum-starved or treated with PMA for 30 min as indicated. Sumoylation and phosphorylation status were determined as in (A, B).
Figure 6
Mapping the determinants in PIASxα for coactivation of Elk-1. (A) Schematic of PIASxα proteins used in this figure. The known PIAS domains (SAP, RING and SUMO binding) are indicated in dark grey boxes. (B) Reporter gene analysis of activities of GAL-Elk(1–428) in the presence or absence of the indicated PIASxα proteins on the GAL-driven E1b promoter-reporter construct in 293 cells. The data are shown relative to GAL-Elk(1–428) in the absence of PIASxα. (C) Western blots probed with Flag (PIASxα) and Gal4 (GAL-Elk) antibodies. Arrows indicate the band corresponding to the protein with the correct size. (D) Co-immunoprecipitation analysis of the indicated PIASxα deletion mutants and cotransfected Elk-1 using an anti-Elk-1 antibody. Arrows represent the locations of the truncated PIASxα proteins. (E) Reporter gene analysis of increasing amounts of the indicated PIASxα proteins (wild type (WT), 125 and 250 ng; 103–572, 500 and 1000 ng; 304–572, 750 and 1500 ng) and the Elk-1 derivative GAL-Elk(205–428) on the GAL-driven E1b promoter-reporter construct in 293 cells. Expression levels of the mutants are shown in the accompanying Western blot. (F, G) Reporter gene analysis of activities of egr-1 promoter-driven luciferase reporter construct in 293 cells. (F) Reporter activity in the absence (white bar) or presence of increasing amounts of WT (light grey bars) or mutant (Δ467–487) (0.1 and 1 μg) (dark grey bars) PIASxα proteins (relative to the basal level of the reporter construct). (G) Reporter activities in the absence (white bar) or presence of WT or mutant (Δ467–487) (dark grey bars) PIASxα proteins in the presence or absence of PMA. The data are shown relative to the basal level of the reporter construct (taken as 1). Luciferase assays are representative of at least two independent experiments (standard errors are shown; _n_=2).
Figure 7
PIASxα upregulates p300 activity. (A, B) Reporter gene analysis of activities of indicated GAL-p300 fusion proteins on the GAL-driven E1b promoter-reporter construct in 293 cells. (A) The activities of the indicated GAL-p300 constructs were tested in the presence or absence of increasing amounts of transfected PIASxα (10, 25 and 100 ng). Data are presented as fold activation of each construct in the absence of PIASxα. (B) Reporter gene analysis of activities of WT GAL-p300 in the presence and absence of PIASxα. The data are shown relative to GAL-p300 in the absence of PIASxα. (C–E) Reporter gene analysis of activities of GAL-Elk(1–428) WT and K2R in the presence and absence of the indicated p300 constructs and/or PIASxα proteins and/or PMA on the GAL-driven E1B promoter-reporter construct in COS7 cells. Data are shown relative to the activity of GAL-Elk(1–428) alone (taken as 1). All samples in panels D and E were stimulated with PMA. (F) Reporter assays were carried out in 293 cells in the presence and absence of siRNA duplexes against PIASxα in serum-free (basal) or in stimulated (PMA) conditions. Luciferase assays are representative of at least two independent experiments (standard errors are shown; _n_=2).
Figure 8
PIASxα affects HDAC-2 binding and histone acetylation levels at Elk-1-regulated promoters. (A–C) Reporter gene analysis of the activities of GAL-Elk-1 constructs on a GAL-driven E1B promoter-reporter construct in 293 cells. (A) The activity of GAL-Elk(205–428) in the presence or absence of PIASxα and/or TSA. Data are shown as luciferase assays relative to GAL-Elk(205–428) alone and as fold induction by TSA. (B) The activities of wild-type (WT) and mutant (K2R) GAL-Elk(1–428) in the presence and absence of PIASxα and/or indicated HDACs. Data are shown as fold induction to the reporter activity relative to each of GAL fusion proteins in the absence of PIASxα (taken as 1). Note the different scales on the axes for WT and K2R versions of Elk-1. (C) The activities of GAL-Elk(1–428) WT and K2R in the presence or absence of RNAi duplexes against GAPDH (−) or the indicated HDACs. Data are shown as fold induction to the reporter activity relative to each of GAL fusion proteins in the presence of GAPDH RNAi duplexes. (D, E) ChIP analysis of HDAC-2, PIASxα (D) and HDAC-6 (E) recruitment to a GAL-driven E1b promoter-reporter construct in 293 cells. The recruitment is monitored following immunoprecipitation (IP) with the indicated antibodies in the presence of the indicated combinations of Gal4 DBD, Gal-Elk(205–428) or Gal-p300 and PIASxα. (F, G) ChIP analysis of HDAC-2 and acetyl-H4 levels on the c-fos promoter. (F) Assays were carried out in 293 cells in the presence of transfected Elk-1 and in the presence or absence of the indicated PIASxα proteins using antisera specific for the indicated proteins. Following immunoprecipitation (IP) of crosslinked lysates, real-time PCR analysis of eluted DNA was performed. All quantitative PCRs were normalised to the input control. Data are shown as relative enrichment for each individual antibody used. (G) ChIP analysis of HeLa cells containing transfected RNAi duplexes against PIASxα using antisera specific for the indicated proteins. (H) Model for the action of PIASxα in facilitating HDAC-2 removal from sumoylated Elk-1. The ERK pathway causes HDAC-2 release and Elk-1 phosphorylation and desumoylation. PIASxα promotes HDAC-2 release and is required for desumoylation. A poised ‘intermediate' sumoylated form of Elk-1 (indicated by ‘?'), which precedes the appearance of a phosphorylated, desumoylated fully active form, is indicated to reflect the molecular role of PIASxα.
Similar articles
- SUMO promotes HDAC-mediated transcriptional repression.
Yang SH, Sharrocks AD. Yang SH, et al. Mol Cell. 2004 Feb 27;13(4):611-7. doi: 10.1016/s1097-2765(04)00060-7. Mol Cell. 2004. PMID: 14992729 - Transforming growth factor-beta-mediated signaling via the p38 MAP kinase pathway activates Smad-dependent transcription through SUMO-1 modification of Smad4.
Ohshima T, Shimotohno K. Ohshima T, et al. J Biol Chem. 2003 Dec 19;278(51):50833-42. doi: 10.1074/jbc.M307533200. Epub 2003 Sep 26. J Biol Chem. 2003. PMID: 14514699 - PIASx is a transcriptional co-repressor of signal transducer and activator of transcription 4.
Arora T, Liu B, He H, Kim J, Murphy TL, Murphy KM, Modlin RL, Shuai K. Arora T, et al. J Biol Chem. 2003 Jun 13;278(24):21327-30. doi: 10.1074/jbc.C300119200. Epub 2003 Apr 25. J Biol Chem. 2003. PMID: 12716907 - SUMO and transcriptional repression: dynamic interactions between the MAP kinase and SUMO pathways.
Yang SH, Jaffray E, Senthinathan B, Hay RT, Sharrocks AD. Yang SH, et al. Cell Cycle. 2003 Nov-Dec;2(6):528-30. doi: 10.4161/cc.2.6.597. Cell Cycle. 2003. PMID: 14504467 Review. - Critical Protein-Protein Interactions Determine the Biological Activity of Elk-1, a Master Regulator of Stimulus-Induced Gene Transcription.
Thiel G, Backes TM, Guethlein LA, Rössler OG. Thiel G, et al. Molecules. 2021 Oct 11;26(20):6125. doi: 10.3390/molecules26206125. Molecules. 2021. PMID: 34684708 Free PMC article. Review.
Cited by
- Emerging roles of SUMO modification in arthritis.
Yan D, Davis FJ, Sharrocks AD, Im HJ. Yan D, et al. Gene. 2010 Oct 15;466(1-2):1-15. doi: 10.1016/j.gene.2010.07.003. Epub 2010 Jul 11. Gene. 2010. PMID: 20627123 Free PMC article. Review. - PIAS1 interacts with FLASH and enhances its co-activation of c-Myb.
Alm-Kristiansen AH, Lorenzo PI, Molværsmyr AK, Matre V, Ledsaak M, Sæther T, Gabrielsen OS. Alm-Kristiansen AH, et al. Mol Cancer. 2011 Feb 21;10:21. doi: 10.1186/1476-4598-10-21. Mol Cancer. 2011. PMID: 21338522 Free PMC article. - Polycomb repressive complex 2 and H3K27me3 cooperate with H3K9 methylation to maintain heterochromatin protein 1α at chromatin.
Boros J, Arnoult N, Stroobant V, Collet JF, Decottignies A. Boros J, et al. Mol Cell Biol. 2014 Oct 1;34(19):3662-74. doi: 10.1128/MCB.00205-14. Epub 2014 Jul 21. Mol Cell Biol. 2014. PMID: 25047840 Free PMC article. - The ETS family transcription factor ELK-1 regulates induction of the cell cycle-regulatory gene p21(Waf1/Cip1) and the BAX gene in sodium arsenite-exposed human keratinocyte HaCaT cells.
Shin SY, Kim CG, Lim Y, Lee YH. Shin SY, et al. J Biol Chem. 2011 Jul 29;286(30):26860-72. doi: 10.1074/jbc.M110.216721. Epub 2011 Jun 3. J Biol Chem. 2011. PMID: 21642427 Free PMC article. - Identification of a new site of sumoylation on Tel (ETV6) uncovers a PIAS-dependent mode of regulating Tel function.
Roukens MG, Alloul-Ramdhani M, Vertegaal AC, Anvarian Z, Balog CI, Deelder AM, Hensbergen PJ, Baker DA. Roukens MG, et al. Mol Cell Biol. 2008 Apr;28(7):2342-57. doi: 10.1128/MCB.01159-07. Epub 2008 Jan 22. Mol Cell Biol. 2008. PMID: 18212042 Free PMC article.
References
- Aravind L, Koonin EV (2000) SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci 25: 112–114 - PubMed
- Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K (1997) Specific inhibition of Stat3 signal transduction by PIAS3. Science 278: 1803–1805 - PubMed
- Gill G (2003) Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr Opin Genet Dev 13: 108–113 - PubMed
- Gill G (2004) SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev 18: 2046–2059 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous