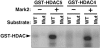An expression screen reveals modulators of class II histone deacetylase phosphorylation - PubMed (original) (raw)
An expression screen reveals modulators of class II histone deacetylase phosphorylation
Shurong Chang et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Class II histone deacetylases (HDACs) repress transcription by associating with a variety of transcription factors and corepressors. Phosphorylation of a set of conserved serine residues in the N-terminal extensions of class II HDACs creates binding sites for 14-3-3 chaperone proteins, which trigger nuclear export of these HDACs, thereby derepressing specific target genes in a signal-dependent manner. To identify intracellular signaling pathways that control phosphorylation of HDAC5, a class II HDAC, we designed a eukaryotic cDNA expression screen in which a GAL4-dependent luciferase reporter was expressed with the DNA-binding domain of GAL4 fused to the N-terminal extension of HDAC5 and the VP16 transcription activation domain fused to 14-3-3. The transfection of COS cells with cDNA expression libraries results in activation of luciferase expression by cDNAs encoding HDAC5 kinases or modulators of such kinases that enable phosphorylated GAL4-HDAC5 to recruit 14-3-3-VP16 with consequent reconstitution of a functional transcriptional complex. Our results reveal a remarkable variety of signaling pathways that converge on the signal-responsive phosphorylation sites in HDAC5, thereby enabling HDAC5 to connect extracellular signals to the genome.
Figures
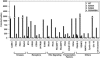
Fig. 1.
Schematic diagram of HDAC5 and the cDNA expression screening strategy. (A) Schematic diagram of HDAC5. The positions of the two signal-responsive serines flanking the nuclear localization sequence (NLS) in the N-terminal extension of HDAC5 are shown. Amino acid positions are indicated. (B) The cDNA expression screen. The N-terminal extension of HDAC5 was fused to the DNA-binding domain of GAL4, and 14-3-3 was fused to the activation domain of VP16. A luciferase reporter controlled by the GAL4 DNA-binding site, referred to as the UAS, is expressed at a basal level in control COS cells. Transfection of COS cells with pools of cDNAs results in the activation of UAS-luciferase expression by pools containing kinases or activators of kinases that phosphorylate the 14-3-3-binding sites in HDAC5, resulting in the recruitment of 14-3-3-VP16 and reconstitution of a transcriptional complex. (C) Results from a transfection assay in a representative 96-well plate are shown. Each well received a pool of ≈50–100 cDNAs as described in B. The UAS-luciferase plasmid was specifically activated in well F2. Sib-selection from this pool identified ET-1 receptor A as the activating cDNA.
Fig. 2.
Activation of UAS-luciferase by expression of cDNAs that promote the association of GAL4-HDAC5 and 14-3-3-VP16. COS cells were transfected with UAS-luciferase and expression plasmids encoding GAL4 fused to the wild-type HDAC5 N-terminal extension or mutants of this region in which serine 259 and/or 498 were mutated to alanines, as indicated, along with 14-3-3-VP16 and expression plasmids for individual activating cDNAs. Mutation of single serines severely impaired activation of UAS-luciferase and mutation of both serines abolished activation.
Fig. 3.
Phosphorylation of HDAC4 and HDAC5 by extracts from Mark2-transfected cells. COS cells were transiently transfected with an empty pcDNA3 expression plasmid (-) or a pcDNA3-Mark2 expression plasmid (+). Cell extracts were prepared and used for in vitro kinase assays with GST–HDAC fusion proteins and [γ-32P]ATP. WT, GST fusion proteins with the wild-type amino acid sequence; Mut, GST fusion proteins in which the signal-responsive serines were mutated to alanines.
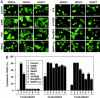
Fig. 4.
Effects of activating cDNAs on nuclear/cytoplasmic distribution of class II HDACs. (A) COS cells were transiently transfected with expression plasmids encoding FLAG-tagged HDACs, as shown in each column, and activators of HDAC5 phosphorylation, as shown in each row. The subcellular distribution of HDACs was determined by immunostaining as described in Materials and Methods. Representative fields are shown. (B) The percentage of cells containing FLAG-tagged HDACs in the cytoplasm was determined by counting at least 100 transfected cells.
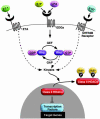
Fig. 5.
Signaling pathways directed at serines 259 and 498 of HDAC5. The ability of class II HDACs to inhibit activity of transcription factors is abolished by phosphorylation at signal-responsive serines, which creates 14-3-3 docking sites and results in the nuclear export of the complex. A variety of inducers of cardiac hypertrophy, including ET-1, sphingosine-1 phosphate, lysophosphatidic acid, serotonin (5-HT), and RhoA signaling, stimulate HDAC kinase activity and consequently derepress HDAC targeted transcription factors. We propose that class II HDACs serve as a nodal point that transmits extracellular and intracellular signals to the genome and controls gene expression.
Similar articles
- Class II HDACs mediate CaMK-dependent signaling to NRSF in ventricular myocytes.
Nakagawa Y, Kuwahara K, Harada M, Takahashi N, Yasuno S, Adachi Y, Kawakami R, Nakanishi M, Tanimoto K, Usami S, Kinoshita H, Saito Y, Nakao K. Nakagawa Y, et al. J Mol Cell Cardiol. 2006 Dec;41(6):1010-22. doi: 10.1016/j.yjmcc.2006.08.010. Epub 2006 Oct 2. J Mol Cell Cardiol. 2006. PMID: 17011572 - Class II histone deacetylases: structure, function, and regulation.
Bertos NR, Wang AH, Yang XJ. Bertos NR, et al. Biochem Cell Biol. 2001;79(3):243-52. Biochem Cell Biol. 2001. PMID: 11467738 Review. - Protein kinase D directly phosphorylates histone deacetylase 5 via a random sequential kinetic mechanism.
Huynh QK, McKinsey TA. Huynh QK, et al. Arch Biochem Biophys. 2006 Jun 15;450(2):141-8. doi: 10.1016/j.abb.2006.02.014. Epub 2006 Mar 9. Arch Biochem Biophys. 2006. PMID: 16584705 - CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy.
Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. Backs J, et al. J Clin Invest. 2006 Jul;116(7):1853-64. doi: 10.1172/JCI27438. Epub 2006 Jun 8. J Clin Invest. 2006. PMID: 16767219 Free PMC article. - Class IIa HDACs - new insights into their functions in physiology and pathology.
Parra M. Parra M. FEBS J. 2015 May;282(9):1736-44. doi: 10.1111/febs.13061. Epub 2014 Oct 27. FEBS J. 2015. PMID: 25244360 Review.
Cited by
- Sumoylation-independent activation of Calcineurin-NFAT-signaling via SUMO2 mediates cardiomyocyte hypertrophy.
Bernt A, Rangrez AY, Eden M, Jungmann A, Katz S, Rohr C, Müller OJ, Katus HA, Sossalla ST, Williams T, Ritter O, Frank D, Frey N. Bernt A, et al. Sci Rep. 2016 Oct 21;6:35758. doi: 10.1038/srep35758. Sci Rep. 2016. PMID: 27767176 Free PMC article. - mef2ca is required in cranial neural crest to effect Endothelin1 signaling in zebrafish.
Miller CT, Swartz ME, Khuu PA, Walker MB, Eberhart JK, Kimmel CB. Miller CT, et al. Dev Biol. 2007 Aug 1;308(1):144-57. doi: 10.1016/j.ydbio.2007.05.018. Epub 2007 May 24. Dev Biol. 2007. PMID: 17574232 Free PMC article. - Regulation of HDAC9 gene expression by MEF2 establishes a negative-feedback loop in the transcriptional circuitry of muscle differentiation.
Haberland M, Arnold MA, McAnally J, Phan D, Kim Y, Olson EN. Haberland M, et al. Mol Cell Biol. 2007 Jan;27(2):518-25. doi: 10.1128/MCB.01415-06. Epub 2006 Nov 13. Mol Cell Biol. 2007. PMID: 17101791 Free PMC article. - Histone deacetylase 5 acquires calcium/calmodulin-dependent kinase II responsiveness by oligomerization with histone deacetylase 4.
Backs J, Backs T, Bezprozvannaya S, McKinsey TA, Olson EN. Backs J, et al. Mol Cell Biol. 2008 May;28(10):3437-45. doi: 10.1128/MCB.01611-07. Epub 2008 Mar 10. Mol Cell Biol. 2008. PMID: 18332106 Free PMC article. - Foxj3 transcriptionally activates Mef2c and regulates adult skeletal muscle fiber type identity.
Alexander MS, Shi X, Voelker KA, Grange RW, Garcia JA, Hammer RE, Garry DJ. Alexander MS, et al. Dev Biol. 2010 Jan 15;337(2):396-404. doi: 10.1016/j.ydbio.2009.11.015. Epub 2009 Nov 13. Dev Biol. 2010. PMID: 19914232 Free PMC article.
References
- Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074-1080. - PubMed
- Grozinger, C. M. & Schreiber, S. L. (2002) Chem. Biol. 9, 3-16. - PubMed
- Verdin, E., Dequiedt, F. & Kasler, H. G. (2003) Trends Genet. 19, 286-293. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases