Conditional transgenic system for mouse aurora a kinase: degradation by the ubiquitin proteasome pathway controls the level of the transgenic protein - PubMed (original) (raw)
Conditional transgenic system for mouse aurora a kinase: degradation by the ubiquitin proteasome pathway controls the level of the transgenic protein
Tomokazu Fukuda et al. Mol Cell Biol. 2005 Jun.
Erratum in
- Correction for Fukuda et al., Conditional Transgenic System for Mouse Aurora A Kinase: Degradation by the Ubiquitin Proteasome Pathway Controls the Level of the Transgenic Protein.
Fukuda T, Mishina Y, Walker MP, DiAugustine RP. Fukuda T, et al. Mol Cell Biol. 2015 Dec;35(23):4094. doi: 10.1128/MCB.00735-15. Mol Cell Biol. 2015. PMID: 26518639 Free PMC article. No abstract available.
Abstract
Aurora A is a mitotic kinase that localizes to centrosomes. Expression of this protein is normally limited to the mitotic stage (G(2)-M) of the cell cycle, whereas human cancer cells frequently exhibit overexpression of Aurora A protein regardless of the cell cycle stage. In the present study, Aurora A transgenic mouse lines were generated with a new conditional expression system (cytomegalovirus immediate early enhancer-chicken beta-actin hybrid promoter-Z-enhanced green fluorescent protein) in order to analyze the function of this protein. Although transcripts for Aurora A were elevated in multiple organs of the transgenic mice, the corresponding protein was not detected in extracts analyzed by immunoblotting. The treatment of transgenic-derived embryonic fibroblasts (MEF) with proteasome inhibitors markedly increased the protein level of transgenic Aurora A, indicating that the transgenic Aurora A protein is readily degraded in normal mouse tissues. Under the exponential growth conditions of MEF cells, transgenic Aurora A was detected within the mitotic stage of the cell cycle and localized to centrosomes. In contrast, the marker of the transgenic promoter (enhanced green fluorescent protein) was continuously expressed throughout the cell cycle, indicating the constitutive transcription of transgenic mRNA. These results indicate that transgenic Aurora A is protected from degradation within G(2)-M but is immediately degraded after translation in the G(1)-S stage of the cell cycle. The findings obtained with this transgenic model and derived cells support that the transition from protection to degradation by the ubiquitin proteasome system at the end of mitosis is an important step in controlling the level of Aurora A protein during the cell cycle.
Figures
FIG. 1.
Strategy for the conditional transgenic CAG-EGFP system. Top, structure of conditional mouse Aurora A transgene using the CAG-Z-EGFP system. CAG, cytomegalovirus immediate early enhancer-chicken beta-actin hybrid promoter; T-lacZ, beta galactosidase with nuclear localization signal; tpA, triple polyadenylation signal; IRES, encephalomyocarditis virus internal ribosome entry site; EGFP, enhanced green fluorescent protein; pA, polyadenylation signal. Left, T-lacZ will be transcribed from the transgenic cassette. Right, Cre recombinase will excise the DNA sequence flanked by loxP sequences. Ribosomes can access the bicistronic mRNA either at the 5′ end to translate the mouse Aurora A or at the IRES to translate the EGFP reporter gene. Parallel protein translation between Aurora A and EGFP proteins should be observed.
FIG. 2.
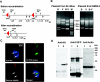
Cre-mediated recombination of the transgenic cassette in Cre-expressing bacteria and the transient expression of Aurora A protein in COS7 cells. (A) The predicted restriction plasmid map for the conditional Aurora A transgenic mouse. B and V indicate the positions for BamHI and EcoRV. The red triangles indicate loxP sequences. The size of each DNA fragment is indicated below. The predicted plasmid maps before and after the recombination are shown. (B) The restriction pattern of DNA fragments of Aurora A transgenic plasmid after the introduction to normal bacteria (XL-1 Blue) or Cre-expressing bacteria (BM25.8). Enzymes used for the digestions are indicated at the top of each lane; BamHI (B), EcoRV (V), and double digestion for BamHI and EcoRV (B-V). Molecular weight markers (M) are expressed in kilobases. Note that the actual restriction pattern of the fragment obtained from BM25.8 matched with that predicted in the map (after recombination in panel A). (C) Transient expression of HA-tagged mouse Aurora A protein in COS7 cells transfected by recombined transgenic constructs. The HA-tagged mouse Aurora A protein is visualized with the antibodies to HA and Alexa 350 (upper left, blue panel). Arrows indicate the position of the centrosome. Centrosomes are visualized by the antibody to gamma-tubulin and Alexa 546 (lower left, red panel). DNA is visualized by the SYTOX-Green (upper right, green panel). The overlap of staining is shown in the merged picture at the lower right. Note that HA antibody shows blue staining at the position of centrosome and cytoplasm. Bar, 5 μm. (D) The Western blot detection of Aurora A protein in the lysate of COS7 cells transfected by the recombined transgenic cassette. Results obtained with the HA, EGFP, and actin antibodies are shown in each column. The specific band obtained with each antibody is indicated by an asterisk. Lane 1, cells transfected by recombined blank transgenic plasmid; lane 2, cells transfected with recombined Aurora A transgenic plasmid. Note that the specific band at 45 kDa with anti-HA was detected only in lane 2 but not in lane 1. The nonspecific band(s) is indicated by NS.
FIG. 3.
LacZ and EGFP detection in the tissues of double transgenic mice (Aurora A/MMTV-Cre). (A) LacZ expression in the tail of a wild-type mouse (upper panel, wild type) and the tail of an Aurora A transgenic mouse (lower panel, Aur A) as detected by X-gal staining. (B) EGFP and LacZ detection in the tail tissue of the Aurora A/MMTV-Cre double transgenic mice and MMTV-Cre mouse. Upper panel, macroscopic observation of tails. Middle panel, EGFP fluorescence detection in double (Aur A/Cre) and single transgenic tails (Cre). Aur A/Cre indicates the Aurora A and MMTV-Cre double transgenic mice. Cre indicates the MMTV-Cre transgenic mouse. The EGFP fluorescence was detected in double transgenic mice (Aur A/Cre) but not in MMTV-Cre mouse (Cre). The EGFP fluorescence was activated upon Cre-mediated excision of floxed T-lacZ and poly(A) sequence. Note the difference in the intensity of EGFP fluorescence among the three tails from double transgenic mice. Lower panel, LacZ expression detected by X-gal staining. Note lacZ expression is detected in the tail tissues of two double transgenic mice that showed weak fluorescence in the middle panel. (C) The detection of EGFP fluorescence in the kidney and brain of double transgenic mouse. Upper panels, macroscopic observation of brain and kidney. Lower panel, EGFP fluorescence intensity in the tissues of double transgenic mice (Aur A/Cre) is much higher than that of MMTV-Cre mouse (Cre). (D) PCR detection of Cre-loxP recombination in the tail tissues of the Aurora A/MMTV-Cre double transgenic mice. The genomic DNA obtained from tails of wild-type, Aurora A transgenic mice, Aurora A/MMTV-Cre double transgenic mice, and MMTV-Cre transgenic mice were subjected to PCR analysis. The corresponding annealing positions of each PCR primer within the transgenic cassette are listed in Fig. 2A. TF41/TF51 is the specific primer set that allows amplification of the unrecombined transgenic allele. TF41/TF91 are the primers that allow the specific amplification of the recombined allele. Note that only the Aurora A/MMTV-Cre double transgenic shows the recombined specific amplification due to the Cre-mediated excision of floxed T-lacZ and poly(A) sequence.
FIG. 4.
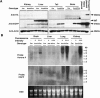
Detection of transgenic Aurora A protein and mRNA in Aurora A, MMTV-Cre double transgenic, and MMTV-Cre mice. (A) The detection of HA-tagged Aurora A, EGFP, and actin in tissues of double transgenic (Aur A/Cre) and MMTV-Cre mice (Cre) by Western blotting. Results obtained from two mice for each genotype are shown in this picture. Protein from COS7 cells transfected with recombined blank vector and recombined transgenic vector was analyzed by immunoblotting as a negative and positive control (transient expression). Note that EGFP expression is observed only in double transgenics but not in MMTV-Cre mice. Note there is no difference in the column of HA between double transgenic (Aur A/Cre) and MMTV-Cre mice (Cre). The IgG band (around 55 kDa) was detected due to the binding of secondary antibody in liver sample. (B) Detection of transgenic mRNA by Northern blot. The results from two double transgenic mice (Aur A/Cre) and one MMTV-Cre mouse (Cre) are shown. The double transgenic mice show different EGFP fluorescence intensities under the microscope even though these two mice are the same genotype (see the fluorescence intensity). Total RNA from brain, liver, lung, and kidney are analyzed. Top, the results obtained with the Aurora A probe. Middle, the results obtained with the EGFP probe. Bottom, ethidium bromide staining of total RNA. Note that the signal derived from the Aurora A probe showed the same expression pattern with that derived from the EGFP probe.
FIG. 5.
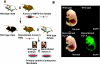
Establishment of mouse embryonic fibroblasts (MEF) containing the recombined Aurora A transgenic cassette. (A) Schematic representation to show how MEF cells that have the recombined Aurora A transgenic cassette were obtained. A wild-type male was bred with a double transgenic female (Aurora A and MMTV-Cre). Due to the Cre expression during oogenesis, a male mouse was obtained that has a recombined Aurora A transgenic cassette at the germ line level. As a result of mating between a recombined Aurora A transgenic male and wild-type females, mouse embryos were obtained that have the recombined Aurora A transgenic cassette throughout the organs. MEF cells were established from the primary culture of embryonic tissues. (B) Detection of the EGFP reporter protein in the mouse transgenic embryo that has the recombined allele. Note that EGFP expression is observed in the recombined Aurora A transgenic mouse embryo.
FIG. 6.
Expression of transgenic Aurora A in MEF cells treated with proteasome inhibitors. Transgenic and wild-type MEF cells were treated with several proteasome inhibitors. Lysates were analyzed by immunoblotting for HA, Aurora A, EGFP, and actin. The results obtained following treatment with (A) MG132, (B) LLnL, (C) cLL, and (D) ALLM are shown. Lysates from COS7 cells transfected with the recombined transgenic vector and empty vector were used as positive and negative controls, respectively (transient expression). Two independent samples were tested for each genotype. Note that an increase in Aurora A transgenic protein was detected in transgenic MEF cells treated with MG132, LLnL, and cLL but not in MEF cells treated with vehicle (dimethylsulfoxide). EGFP expression in MEF cells derived from the recombined transgenic showed no difference whether treated with or without inhibitors.
FIG. 7.
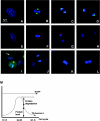
Detection of G2-M-dependent expression of native and transgenic Aurora A protein in wild-type and transgenic MEF cells. (A to D) Detection of native Aurora A protein at the following stages of cell cycle: (A) interphase, (B) metaphase, (C) late anaphase, and (D) cytokinesis. Native Aurora A is visualized by green fluorescence (Alexa 488). The chromosome is visualized by blue fluorescence (4_N_,6_N_-diamidino-2-phenylindole). (E to H) Detection of transgenic Aurora A protein in transgenic-derived MEF cells at the following stages of cell cycle: (E) interphase, (F) metaphase, (G) late anaphase, and (H) cytokinesis. Transgenic Aurora A protein is visualized by red fluorescence (Alexa 546). (I) The detection of EGFP reporter protein in cycling MEF cells. The mitotic stage of cell division is denoted by an asterisk. Note that the fluorescence intensity of the EGFP marker protein during cell division appears similar to that observed when the cells are in interphase (arrow). (J) Detection of EGFP in wild-type MEF cells. (K) Control staining of native Aurora A (IgG control). (L) Control staining of HA antibody in wild-type MEF cells. (M) Model for the cell cycle dependent expression of transgenic Aurora A protein. The longitudinal axis represents the level of protein expression. The horizontal axis indicates the stage of the cell cycle. The transgenic promoter is active throughout the cell cycle (as represented by EGFP). Aurora A is protected from the degradation during the G2-M of the cell cycle but is degraded during G1-S of the cell cycle.
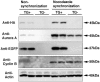
FIG. 8.
Changes in protein level of native and transgenic Aurora A in nocodazole-synchronized MEF cells. Wild-type and transgenic MEF cells were treated with vehicle (dimethyl sulfoxide) or nocodazole. Lysates were analyzed by immunoblotting for HA, Aurora A, EGFP, cyclin B, and actin. Two independent samples were tested for each genotype. A comparable increase in cyclin B was observed in transgenic (TG+) and wild-type (TG−) cells exposed to nocodazole. Note that the signal of transgenic Aurora A (anti-HA) increased in the TG+ cells after the nocodazole treatment and that the level of total Aurora A in these cells was at least twofold greater than that of wild-type cells. By contrast, a strong signal for EGFP was detected in the transgenic cells with and without nocodazole treatment.
Similar articles
- Mechanism of Aurora-B degradation and its dependency on intact KEN and A-boxes: identification of an aneuploidy-promoting property.
Nguyen HG, Chinnappan D, Urano T, Ravid K. Nguyen HG, et al. Mol Cell Biol. 2005 Jun;25(12):4977-92. doi: 10.1128/MCB.25.12.4977-4992.2005. Mol Cell Biol. 2005. PMID: 15923616 Free PMC article. - Aurora-A kinase interacting protein 1 (AURKAIP1) promotes Aurora-A degradation through an alternative ubiquitin-independent pathway.
Lim SK, Gopalan G. Lim SK, et al. Biochem J. 2007 Apr 1;403(1):119-27. doi: 10.1042/BJ20061272. Biochem J. 2007. PMID: 17125467 Free PMC article. - Aurora-A overexpression in mouse liver causes p53-dependent premitotic arrest during liver regeneration.
Li CC, Chu HY, Yang CW, Chou CK, Tsai TF. Li CC, et al. Mol Cancer Res. 2009 May;7(5):678-88. doi: 10.1158/1541-7786.MCR-08-0483. Epub 2009 May 12. Mol Cancer Res. 2009. PMID: 19435814 - [Visualizing spatiotemporal dynamics of multicellular cell-cycle progression].
Sakaue-Sawano A, Miyawaki A. Sakaue-Sawano A, et al. Seikagaku. 2012 Jan;84(1):47-52. Seikagaku. 2012. PMID: 22416473 Review. Japanese. No abstract available. - The Non-Canonical Role of Aurora-A in DNA Replication.
Tsunematsu T, Arakaki R, Yamada A, Ishimaru N, Kudo Y. Tsunematsu T, et al. Front Oncol. 2015 Aug 25;5:187. doi: 10.3389/fonc.2015.00187. eCollection 2015. Front Oncol. 2015. PMID: 26380219 Free PMC article. Review.
Cited by
- Generation of a new mouse line with conditionally activated signaling through the BMP receptor, ACVR1: A tool to characterize pleiotropic roles of BMP functions.
Yang J, Toda Nakamura M, Hallett SA, Ueharu H, Zhang H, Kelley K, Fukuda T, Komatsu Y, Mishina Y. Yang J, et al. Genesis. 2021 Jun;59(5-6):e23419. doi: 10.1002/dvg.23419. Epub 2021 Apr 14. Genesis. 2021. PMID: 33851764 Free PMC article. - The anti-proliferative effects of the CHFR depend on the forkhead associated domain, but not E3 ligase activity mediated by ring finger domain.
Fukuda T, Kondo Y, Nakagama H. Fukuda T, et al. PLoS One. 2008 Mar 12;3(3):e1776. doi: 10.1371/journal.pone.0001776. PLoS One. 2008. PMID: 18335050 Free PMC article. - A FRET biosensor reveals spatiotemporal activation and functions of aurora kinase A in living cells.
Bertolin G, Sizaire F, Herbomel G, Reboutier D, Prigent C, Tramier M. Bertolin G, et al. Nat Commun. 2016 Sep 14;7:12674. doi: 10.1038/ncomms12674. Nat Commun. 2016. PMID: 27624869 Free PMC article. - A genetic variant of Aurora kinase A promotes genomic instability leading to highly malignant skin tumors.
Torchia EC, Chen Y, Sheng H, Katayama H, Fitzpatrick J, Brinkley WR, Caulin C, Sen S, Roop DR. Torchia EC, et al. Cancer Res. 2009 Sep 15;69(18):7207-15. doi: 10.1158/0008-5472.CAN-09-1059. Epub 2009 Sep 8. Cancer Res. 2009. PMID: 19738056 Free PMC article. - Rapid calcium-dependent activation of Aurora-A kinase.
Plotnikova OV, Pugacheva EN, Dunbrack RL, Golemis EA. Plotnikova OV, et al. Nat Commun. 2010 Sep 7;1(6):64. doi: 10.1038/ncomms1061. Nat Commun. 2010. PMID: 20842194 Free PMC article.
References
- Anand, S., S. Penrhyn-Lowe, and A. R. Venkitaraman. 2003. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 3:51-62. - PubMed
- Bischoff, J. R., L. Anderson, Y. Zhu, K. Mossie, L. Ng, B. Souza, B. Schryver, P. Flanagan, F. Clairvoyant, C. Ginther, C. S. Chan, M. Novotny, D. J. Slamon, and G. D. Plowman. 1998. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17:3052-3065. - PMC - PubMed
- Bischoff, J. R., and G. D. Plowman. 1999. The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 9:454-459. - PubMed
- Crane, R., A. Kloepfer, and J. V. Ruderman. 2004. Requirements for the destruction of human Aurora-A. J. Cell Sci. 117:5975-5983. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases