Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer - PubMed (original) (raw)
Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer
Gennadi V Glinsky et al. J Clin Invest. 2005 Jun.
Abstract
Activation in transformed cells of normal stem cells' self-renewal pathways might contribute to the survival life cycle of cancer stem cells and promote tumor progression. The BMI-1 oncogene-driven gene expression pathway is essential for the self-renewal of hematopoietic and neural stem cells. We applied a mouse/human comparative translational genomics approach to identify an 11-gene signature that consistently displays a stem cell-resembling expression profile in distant metastatic lesions as revealed by the analysis of metastases and primary tumors from a transgenic mouse model of prostate cancer and cancer patients. To further validate these results, we examined the prognostic power of the 11-gene signature in several independent therapy-outcome sets of clinical samples obtained from 1,153 cancer patients diagnosed with 11 different types of cancer, including 5 epithelial malignancies (prostate, breast, lung, ovarian, and bladder cancers) and 5 nonepithelial malignancies (lymphoma, mesothelioma, medulloblastoma, glioma, and acute myeloid leukemia). Kaplan-Meier analysis demonstrated that a stem cell-like expression profile of the 11-gene signature in primary tumors is a consistent powerful predictor of a short interval to disease recurrence, distant metastasis, and death after therapy in cancer patients diagnosed with 11 distinct types of cancer. These data suggest the presence of a conserved BMI-1-driven pathway, which is similarly engaged in both normal stem cells and a highly malignant subset of human cancers diagnosed in a wide range of organs and uniformly exhibiting a marked propensity toward metastatic dissemination as well as a high probability of unfavorable therapy outcome.
Figures
Figure 1
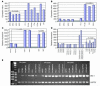
Microarray (A–D) and RT-PCR (E) analyses reveal increased expression of BMI-1 mRNA in multiple human prostate cancer cell lines established from metastatic tumors (PC-3, LNCap, DuCap, VCap, etc.) compared with normal human prostate epithelial cells (NPEC) (A and E); in xenograft-derived human prostate cancer cell line variants (PC-3M, PC-3MLN4, PC-3MPro4) compared with the plastic-maintained parental cells (PC-3) (B); in highly metastatic human prostate carcinoma xenografts (PC-3MLN4) compared with the less metastatic parental counterparts (PC-3) growing orthotopically in nude mice (C); in lymph node metastases of human prostate cancer growing in the prostate of nude mice (MET) (C); and in invasive primary prostate tumors and distant metastatic lesions in the TRAMP transgenic mouse model of prostate cancer (D). Prostate tissues from age-matched wild-type C57BL/6 mice served as control samples in Figure 1D. The numbers 4, 5, and 7 indicate the age of TRAMP mice (in months). Each sample represents a pool of tissues from 3–5 mice. P values were obtained using a 2-tailed t test. LN3, LNCapLN3; LN4, PC-3MLN4; PRO4, PC-3MPRO4; PRO5, LNCapPRO5; SV, seminal vesicles; X, human xenograft tumors in nude mice.
Figure 2
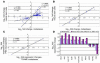
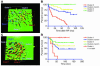
Distant metastatic lesions in the TRAMP transgenic mouse model of prostate cancer exhibit stem cell–like expression signatures of the BMI-1 pathway. Transcripts differentially regulated in distant metastatic lesions of 6-month-old TRAMP mice (MTTS signature) were compared with the BMI-1–regulated genes in neural stem cells (3) in search of intersection of lists. (A) Expression profiles and the corresponding Pearson correlation coefficient for 199 genes (141 upregulated and 58 downregulated) comprising concordant differentially regulated sets of transcripts in metastatic TRAMP samples and PNS neurospheres are shown. Small gene expression signatures comprising transcripts with a high level of expression correlation in metastatic cancer cells and stem cells (the selection threshold for small signatures was arbitrarily set at Pearson correlation coefficients greater than 0.95) were selected from large concordant sets. The reduction in the signature transcript number was terminated when further elimination of a transcript did not increase the value of the Pearson correlation coefficient. Using this approach, a single candidate prognostic gene expression signature was selected for each binary intersection of the MTTS signature and parent stem cell signatures (Figure 3). Consecutive steps of selection from the 199-gene concordant set of a subset of 20 genes (A and B) and a small MTTS/PNS 11-gene signature (C and D) are shown. In D, r = 0.9897, P < 0.0001 between gene groups, and n = 11 per group. Complete lists of genes and corresponding concordant subsets are shown in Supplemental Table 2. See text, Figure 3, and Table 3 for details.
Figure 3
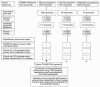
Sequential analytical steps used for identification, selection, and validation of the 11-gene death-from-cancer signature. The figure shows an overview of the approach used for the development and validation of a cancer survival predictor based on gene expression monitoring.
Figure 4
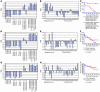
Selection of the best-performing small signature based on evaluation of the metastatic-phenotype-discrimination performance and therapy-outcome prediction power of candidate prognostic signatures. Expression profiles of the 3 small signatures (11-gene MTTS/PNS signature, A–C; 11-gene MTTS/CNS signature, D–F; and 14-gene MTTS/PNS/CNS signature, G–I) were evaluated in metastatic lesions at multiple distant target organs and primary prostate carcinomas in the TRAMP transgenic mouse model of prostate cancer (A, D, and G) and prostate cancer patients (B, E, and H) for presence of a stem cell–like expression profile. (B, E, and H) Data from the analysis of 9 distant metastatic lesions and 23 primary human prostate carcinoma samples. (C, F, and I) Kaplan-Meier analysis of the probability that patients would remain disease-free among 21 prostate cancer patients constituting clinical outcome set 1, according to whether they had a good-prognosis or a poor-prognosis signature as defined by the expression profiles of the small prognostic signatures. The y axes in A, B, D, E, G, and H show the SPAI values in corresponding metastatic and primary tumor samples (see Methods for a description of SPAI definition and calculation). CI, confidence interval.
Figure 5
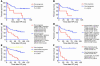
TRN analysis within the mRNA abundance space of genes constituting the 11-gene MTTS/PNS signature reveals clustering patterns, among prostate cancer (A and B) and breast cancer (C and D) patients, that are associated with distinct frequencies of therapy failure (A and B) and differing probability of disease-free survival after therapy (C and D). A TRN clustering algorithm was applied to the 79 samples (A) constituting prostate cancer therapy outcome set 2 and the 97 samples (C) constituting the breast cancer therapy outcome set. Kaplan-Meier analysis (B and D) was applied to subgroups of patients defined by the TRN clustering algorithm as shown in A and C.
Figure 6
Classification of prostate cancer patients into subgroups with distinct therapy outcome based on expression profile of the 11-gene MTTS/PNS signature. (A–C) Kaplan-Meier analysis of the probability that patients would remain disease-free among 79 prostate cancer patients constituting clinical outcome set 2, according to whether they had a good-prognosis or a poor-prognosis signature as defined by the expression profiles of the 11-gene MTTS/PNS signature. The patients’ stratification cutoff value of 0.4 was defined in the training set of 40 patients (19 poor prognosis and 21 good prognosis; A), validated in a test set of 39 patients (18 poor prognosis and 21 good prognosis; B) and confirmed in an entire cohort of 79 patients (C). (D) Kaplan-Meier survival curves for distinct subgroups of prostate cancer patients diagnosed with early-stage disease (stages 1C and 2A). (E) Kaplan-Meier survival curves for 79 prostate cancer patients stratified into distinct subgroups using a weighted survival predictor score algorithm. (F) Kaplan-Meier survival curves for 20 prostate cancer patients stratified into distinct subgroups using Q-RT-PCR assay of the 11-gene signature.
Figure 7
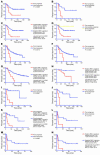
Classification of patients diagnosed with 4 different types of epithelial cancer into subgroups with distinct therapy outcome based on expression profile of the 11-gene MTTS/PNS signa-ture. Kaplan-Meier analysis of the probability that patients would remain metastasis-free (for the breast cancer group) or survive after therapy (for the other groups) among 97 early-stage breast cancer patients (A–D), 125 lung adenocarcinoma patients of all stages (E–G), 35 lung adenocarcinoma patients diagnosed with stage 1A disease (H), 37 ovarian cancer patients of all stages (I–K), and 31 bladder cancer patients (L–N), according to whe-ther they had a good-prognosis or a poor-prognosis signature as defined by the expression profiles of the 11-gene MTTS/PNS signature. For each type of cancer, the patient’s stratification cutoff value was defined in the training set, validated in a test set, and confirmed in an entire cohort. D and I–K show the Kaplan-Meier survival curves for 97 breast cancer patients and 37 ovarian cancer patients, respectively, stratified into distinct subgroups using a weighted survival predictor score algorithm.
Figure 8
Classification of cancer patients diagnosed with different types of nonepithelial malignancies into subgroups with distinct therapy outcome based on expression profile of the 11-gene MTTS/PNS signature. Kaplan-Meier survival analysis of the probability of a therapy failure in cancer patients diagnosed with different types of nonepithelial cancers and having distinct expression profiles of the 11-gene MTTS/PNS signature is shown. Data from lymphoma patients (A), malignant glioma patients (B), mesothelioma patients (C), medulloblastoma patients (D), mantle cell lymphoma patients (E), and acute myeloid leukemia patients (F) are shown.
Comment in
- Stem cell-ness: a "magic marker" for cancer.
Lahad JP, Mills GB, Coombes KR. Lahad JP, et al. J Clin Invest. 2005 Jun;115(6):1463-7. doi: 10.1172/JCI25455. J Clin Invest. 2005. PMID: 15931383 Free PMC article.
Similar articles
- Essential role for activation of the Polycomb group (PcG) protein chromatin silencing pathway in metastatic prostate cancer.
Berezovska OP, Glinskii AB, Yang Z, Li XM, Hoffman RM, Glinsky GV. Berezovska OP, et al. Cell Cycle. 2006 Aug;5(16):1886-901. doi: 10.4161/cc.5.16.3222. Epub 2006 Aug 15. Cell Cycle. 2006. PMID: 16963837 - Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells.
Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS. Liu S, et al. Cancer Res. 2006 Jun 15;66(12):6063-71. doi: 10.1158/0008-5472.CAN-06-0054. Cancer Res. 2006. PMID: 16778178 Free PMC article. - Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences.
Siddique HR, Saleem M. Siddique HR, et al. Stem Cells. 2012 Mar;30(3):372-8. doi: 10.1002/stem.1035. Stem Cells. 2012. PMID: 22252887 Review. - Stem cell divisions controlled by the proto-oncogene BMI-1.
Grinstein E, Mahotka C. Grinstein E, et al. J Stem Cells. 2009;4(3):141-6. J Stem Cells. 2009. PMID: 20232599 Review.
Cited by
- Nuclear-encoded cytochrome c oxidase subunit 4 regulates BMI1 expression and determines proliferative capacity of high-grade gliomas.
Oliva CR, Markert T, Gillespie GY, Griguer CE. Oliva CR, et al. Oncotarget. 2015 Feb 28;6(6):4330-44. doi: 10.18632/oncotarget.3015. Oncotarget. 2015. PMID: 25726526 Free PMC article. - GLI1 regulates a novel neuropilin-2/α6β1 integrin based autocrine pathway that contributes to breast cancer initiation.
Goel HL, Pursell B, Chang C, Shaw LM, Mao J, Simin K, Kumar P, Vander Kooi CW, Shultz LD, Greiner DL, Norum JH, Toftgard R, Kuperwasser C, Mercurio AM. Goel HL, et al. EMBO Mol Med. 2013 Apr;5(4):488-508. doi: 10.1002/emmm.201202078. Epub 2013 Feb 21. EMBO Mol Med. 2013. PMID: 23436775 Free PMC article. - BMI1 polycomb group protein acts as a master switch for growth and death of tumor cells: regulates TCF4-transcriptional factor-induced BCL2 signaling.
Siddique HR, Parray A, Tarapore RS, Wang L, Mukhtar H, Karnes RJ, Deng Y, Konety BR, Saleem M. Siddique HR, et al. PLoS One. 2013 May 6;8(5):e60664. doi: 10.1371/journal.pone.0060664. Print 2013. PLoS One. 2013. PMID: 23671559 Free PMC article. - B-cell-specific Moloney murine leukemia virus integration site 1: potential stratification factor and therapeutic target for epithelial ovarian cancer.
Zhao Q, Gui T, Qian Q, Li L, Shen K. Zhao Q, et al. Onco Targets Ther. 2016 Aug 22;9:5203-8. doi: 10.2147/OTT.S109443. eCollection 2016. Onco Targets Ther. 2016. PMID: 27578986 Free PMC article. Review. - Overexpression of ubiquitin-specific protease 22 predicts poor survival in patients with early-stage non-small cell lung cancer.
Ning J, Zhang J, Liu W, Lang Y, Xue Y, Xu S. Ning J, et al. Eur J Histochem. 2012 Nov 30;56(4):e46. doi: 10.4081/ejh.2012.e46. Eur J Histochem. 2012. PMID: 23361242 Free PMC article.
References
- Lessard J, Sauvageau G. BMI-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. - PubMed
- Park I-K, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. - PubMed
- Dick JE. Self-renewal writ in blood. Nature. 2003;423:231–233. - PubMed
- Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat. Rev. Cancer. 2003;3:895–902. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases