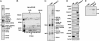Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration - PubMed (original) (raw)
Comparative Study
. 2005 Jun 14;102(24):8472-7.
doi: 10.1073/pnas.0503505102. Epub 2005 Jun 2.
Affiliations
- PMID: 15932940
- PMCID: PMC1150862
- DOI: 10.1073/pnas.0503505102
Comparative Study
Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration
Vikas B Palhan et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Spinocerebellar ataxia type 7 (SCA7) is characterized by cone-rod dystrophy retinal degeneration and is caused by a polyglutamine [poly(Q)] expansion within ataxin-7, a protein of previously unknown function. Here, we report that ataxin-7 is an integral component of the mammalian STAGA (SPT3-TAF9-ADA-GCN5 acetyltransferase) transcription coactivator complex, interacts directly with the GCN5 histone acetyltransferase component of STAGA, and mediates a direct interaction of STAGA with the CRX (cone-rod homeobox) transactivator of photoreceptor genes. Consistent with these results, chromatin immunoprecipitation assays document retinal-specific association of CRX, GCN5, and acetylated histone H3 with CRX target genes. RNA interference studies also implicate ataxin-7 and GCN5 in CRX-dependent gene activation, and histone deacetylase inhibitors restore the compromised expression of a CRX target gene in an ataxin-7-deficient background. Significantly, in relation to SCA7, poly(Q)-expanded ataxin-7 gets incorporated into STAGA and, in a dominant-negative manner, inhibits the nucleosomal histone acetylation function of STAGA GCN5 both in vitro and, based on chromatin immunoprecipitation assays, in SCA7 transgenic mice. These results suggest that the normal function of a poly(Q) disease protein may intersect with its pathogenic mechanism, an observation with significant implications for the molecular basis of all poly(Q) disorders and ultimately for their treatment.
Figures
Fig. 1.
Ataxin-7 is an integral subunit of the STAGA complex. (A) SDS/PAGE/silver stain analysis of affinity-purified STAGA from FLAG.HA-SPT3 HeLa cells. The p110 band was identified as ataxin-7 by tandem MS analysis. Asterisks indicate nonspecific bands. (B) Immunoblot of Sephacryl S-300 fractions after chromatography of HeLa S3 NE. (C) SDS/PAGE/silver stain analysis of control (IgG resin) or anti-ataxin-7 affinity-purified STAGA from Y-79 cell NE. (D) Immunoblot of IPs from C with indicated antibodies. Y-79 NE served as positive control. (E) Fluorogram of HAT assay of IPs from C, using free histones as substrate. Recombinant PCAF served as a positive control.
Fig. 2.
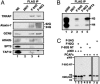
Poly(Q)-expanded ataxin-7 is incorporated into STAGA and inactivates its HAT function in a dominant-negative manner. (A) Immunoblot of anti-FLAG IPs of NE from control 293T cells (lane 2) or from 293T lines stably expressing FLAG-ataxin-7–24Q (lane 3) or FLAG-ataxin-7–92Q (lane 4). 293T NE served as positive control (lane 1). (B) Fluorogram of HAT assays using an oligonucleosome substrate on IPs from A, a highly concentrated STAGA complex (lane 5), and recombinant p300 (lane 1). (C) Fluorogram of solution HAT assays with an oligonucleosome substrate and purified proteins FLAG-ataxin-7–24Q/-92Q, FLAG-ataxin-7–92Q NT, His6-p300, and affinity-purified PCAF complex added as indicated (top).
Fig. 3.
CRX interacts with STAGA through ataxin-7. (A) SDS/PAGE/fluorographic analysis of interactions of 35S-labeled in vitro_-translated proteins (LUC, CRX, and CRX-Mutant) with IPs from Fig. 1_C. Additions were as indicated (top), and inputs (2%) are shown in lanes 1, 4, and 7. (B) SDS/PAGE/fluorographic analysis of interactions of 35S-labeled _in vitro_-translated proteins (GCN5-S and GCN5-L) with FLAG-CRX in the presence and absence of Myc-ataxin-7–92Q (Myc-Atx7). Similar results were obtained with Myc-ataxin-7–10Q (data not shown). Additions were as indicated (top). Corresponding empty vector controls are represented by “-.” Inputs (2%) are shown in lanes 7 and 8. (C) ChIP analysis of acetylated histone H3 and GCN5 on CRX-target genes in mouse retina vs. liver tissues. PCR primers covered the regulatory regions indicated in parentheses. M-opsin, M-cone opsin; LCR, locus control region of M-cone opsin; S-opsin, S-cone opsin; IRBP, interphotoreceptor-binding protein. Input (pre-IP), + and - antibody (Ab), and mock (no DNA) lanes are marked. Similar results were obtained by using an isotype-matched antibody (rabbit IgG, Santa Cruz Biotechnology) as with the no antibody control.
Fig. 4.
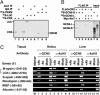
CRX/NRL-mediated activation of the rhodopsin promoter depends on a functional STAGA complex. (A and B) Immunoblot of whole-cell extracts from HEK293 cells transfected with siRNAs as indicated (top). (C) Effects of ataxin-7, GCN5, and p300 siRNAs on expression of a transfected rhodopsin promoter-driven LUC reporter in HEK293 cells. LUC and nonspecific control (C1) siRNAs served as positive and negative controls, respectively. The activation observed from mock-transfected HEK293 cells was arbitrarily set as 100%. (D) HDAC inhibitor-mediated recovery of ataxin-7 siRNA-compromised rhodopsin-promoter activity in transfected HEK293 cells. Control siRNA (C1) and ataxin-7 siRNA transfected cells (as in C) were treated with suberoylanilide hydroxamic acid and butyrate (HDAC inhibitors) as indicated. Error bars represent mean plus standard deviation in C and D from three independent experiments.
Fig. 5.
HAT activity alterations in SCA7 result from a dominant-negative effect on the STAGA complex. ChIP assays on retinas from SCA7–92Q transgenic mice and their nontransgenic WT littermates reveal differences in occupancy of CRX target and nontarget gene regulatory regions. Quantitation of the occupancy differences is provided in Table 1. Brackets group together the enhancer/promoter pair regulating each gene. On the far right, previously reported gene expression alterations for this SCA7 mouse model of retinal degeneration are as indicated (17). SER, S-cone opsin enhancer region; RER, rhodopsin enhancer region; Arr3, cone arrestin 3; IER, IRBP enhancer region. See the legend of Fig. 3_C_ for other definitions. A 100-bp DNA ladder was used as a size marker.
Fig. 6.
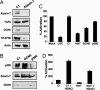
Mutant ataxin-7 effects on GCN5 function. (A) Retinal sections from 13-week-old SCA7–92Q transgenic mice (92Q), SCA7–24Q (24Q), and nontransgenic littermates (WT) were immunostained with an anti-GCN5 antibody (green). Although confocal images reveal intense immunostaining of photoreceptor nuclei (ONL) and bipolar/interneuron nuclei (INL) in the 92Q mice, no appreciable GCN5 immunostaining is detectable in the 24Q or nontransgenic mice. OS, outer segments; IS, inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. (Scale bar, 100 μm.) (B) Models indicating (i) normal function of WT ataxin-7 (Atx7), as an integral GCN5-interacting subunit of STAGA, in facilitating STAGA recruitment and GCN5-mediated histone H3 acetylation through interactions with a promoter-bound transcription factor (TF) such as CRX (Left), and (ii) dominant-negative inhibition of GCN5 function (histone acetylation) within promoter-bound STAGA by poly(Q)-expanded ataxin-7 (Atx7–92Q) after interactions with WT ataxin-7-containing STAGA (Center) or after incorporation into STAGA in place of WT ataxin-7 resulting in reduced incorporation of ADA2b, SPT3, and TAF12 (Right).
Similar articles
- Both normal and polyglutamine- expanded ataxin-7 are components of TFTC-type GCN5 histone acetyltransferase- containing complexes.
Helmlinger D, Hardy S, Eberlin A, Devys D, Tora L. Helmlinger D, et al. Biochem Soc Symp. 2006;(73):155-63. doi: 10.1042/bss0730155. Biochem Soc Symp. 2006. PMID: 16626296 - Interference of Crx-dependent transcription by ataxin-7 involves interaction between the glutamine regions and requires the ataxin-7 carboxy-terminal region for nuclear localization.
Chen S, Peng GH, Wang X, Smith AC, Grote SK, Sopher BL, La Spada AR. Chen S, et al. Hum Mol Genet. 2004 Jan 1;13(1):53-67. doi: 10.1093/hmg/ddh005. Epub 2003 Nov 12. Hum Mol Genet. 2004. PMID: 14613968 - Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes.
Helmlinger D, Hardy S, Sasorith S, Klein F, Robert F, Weber C, Miguet L, Potier N, Van-Dorsselaer A, Wurtz JM, Mandel JL, Tora L, Devys D. Helmlinger D, et al. Hum Mol Genet. 2004 Jun 15;13(12):1257-65. doi: 10.1093/hmg/ddh139. Epub 2004 Apr 28. Hum Mol Genet. 2004. PMID: 15115762 - Transcriptional alterations and chromatin remodeling in polyglutamine diseases.
Helmlinger D, Tora L, Devys D. Helmlinger D, et al. Trends Genet. 2006 Oct;22(10):562-70. doi: 10.1016/j.tig.2006.07.010. Epub 2006 Sep 5. Trends Genet. 2006. PMID: 16911843 Review. - Molecular pathogenesis and cellular pathology of spinocerebellar ataxia type 7 neurodegeneration.
Garden GA, La Spada AR. Garden GA, et al. Cerebellum. 2008;7(2):138-49. doi: 10.1007/s12311-008-0027-y. Cerebellum. 2008. PMID: 18418675 Free PMC article. Review.
Cited by
- Respiratory neuropathology in spinocerebellar ataxia type 7.
Biswas DD, Shi Y, El Haddad L, Sethi R, Huston M, Kehoe S, Scarrow ER, Strickland LM, Pucci LA, Dhindsa JS, Hunanyan A, La Spada AR, ElMallah MK. Biswas DD, et al. JCI Insight. 2024 Jul 18;9(18):e170444. doi: 10.1172/jci.insight.170444. JCI Insight. 2024. PMID: 39053472 Free PMC article. - Polyglutamine (PolyQ) Diseases: Navigating the Landscape of Neurodegeneration.
Tenchov R, Sasso JM, Zhou QA. Tenchov R, et al. ACS Chem Neurosci. 2024 Aug 7;15(15):2665-2694. doi: 10.1021/acschemneuro.4c00184. Epub 2024 Jul 12. ACS Chem Neurosci. 2024. PMID: 38996083 Free PMC article. Review. - Antibody-assisted selective isolation of Purkinje cell nuclei from mouse cerebellar tissue.
Bartelt LC, Fakhri M, Adamek G, Trybus M, Samelak-Czajka A, Jackowiak P, Fiszer A, Lowe CB, La Spada AR, Switonski PM. Bartelt LC, et al. Cell Rep Methods. 2024 Jul 15;4(7):100816. doi: 10.1016/j.crmeth.2024.100816. Epub 2024 Jul 8. Cell Rep Methods. 2024. PMID: 38981474 Free PMC article. - Hereditary Ataxias: From Bench to Clinic, Where Do We Stand?
Pilotto F, Del Bondio A, Puccio H. Pilotto F, et al. Cells. 2024 Feb 9;13(4):319. doi: 10.3390/cells13040319. Cells. 2024. PMID: 38391932 Free PMC article. Review. - UPS writes a new saga of SAGA.
Barman P, Chakraborty P, Bhaumik R, Bhaumik SR. Barman P, et al. Biochim Biophys Acta Gene Regul Mech. 2023 Dec;1866(4):194981. doi: 10.1016/j.bbagrm.2023.194981. Epub 2023 Aug 30. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 37657588 Review.
References
- Zoghbi, H. Y. & Orr, H. T. (2000) Annu. Rev. Neurosci. 23, 217-247. - PubMed
- Grote, S. K. & La Spada, A. R. (2003) Cytogenet. Genome Res. 100, 164-174. - PubMed
- David, G., Abbas, N., Stevanin, G., Durr, A., Yvert, G., Cancel, G., Weber, C., Imbert, G., Saudou, F., Antoniou, E., et al. (1997) Nat. Genet. 17, 65-70. - PubMed
- Feany, M. B. & La Spada, A. R. (2003) Neuron 40, 1-2. - PubMed
- Panov, A. V., Gutekunst, C. A., Leavitt, B. R., Hayden, M. R., Burke, J. R., Strittmatter, W. J. & Greenamyre, J. T. (2002) Nat. Neurosci. 5, 731-736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- EY14061/EY/NEI NIH HHS/United States
- P30 EY002687/EY/NEI NIH HHS/United States
- EY12543/EY/NEI NIH HHS/United States
- R01 EY012543/EY/NEI NIH HHS/United States
- P30 EY001730/EY/NEI NIH HHS/United States
- EY02687/EY/NEI NIH HHS/United States
- R01 EY014061/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials