oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes - PubMed (original) (raw)
oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes
Shannan J Ho Sui et al. Nucleic Acids Res. 2005.
Abstract
Targeted transcript profiling studies can identify sets of co-expressed genes; however, identification of the underlying functional mechanism(s) is a significant challenge. Established methods for the analysis of gene annotations, particularly those based on the Gene Ontology, can identify functional linkages between genes. Similar methods for the identification of over-represented transcription factor binding sites (TFBSs) have been successful in yeast, but extension to human genomics has largely proved ineffective. Creation of a system for the efficient identification of common regulatory mechanisms in a subset of co-expressed human genes promises to break a roadblock in functional genomics research. We have developed an integrated system that searches for evidence of co-regulation by one or more transcription factors (TFs). oPOSSUM combines a pre-computed database of conserved TFBSs in human and mouse promoters with statistical methods for identification of sites over-represented in a set of co-expressed genes. The algorithm successfully identified mediating TFs in control sets of tissue-specific genes and in sets of co-expressed genes from three transcript profiling studies. Simulation studies indicate that oPOSSUM produces few false positives using empirically defined thresholds and can tolerate up to 50% noise in a set of co-expressed genes.
Figures
Figure 1
The oPOSSUM system for identifying over-represented TFBSs in sets of co-expressed genes. The system is built upon a database of conserved TFBSs for human–mouse orthologs, derived from an analysis pipeline that combines phylogenetic footprinting with TFBS identification using the JASPAR library of PSSMs. Given a set of human or mouse genes, the pipeline (1) retrieves the genomic DNA sequence for the human and mouse genes plus 5000 bp of upstream sequence, (2) performs an alignment of the orthologous sequences and extracts non-coding DNA subsequences that are conserved above a predefined threshold, (3) searches the subsequences for matches to TFBS profiles contained in JASPAR and (4) stores the results in the oPOSSUM database. Upon querying the web-based interface with a list of co-expressed genes, oPOSSUM retrieves the TFBS counts for each gene in the list and computes two statistics (_Z_-score, Fisher exact test) to measure over-representation of TFBSs in the set relative to a background comprising all genes in the oPOSSUM database.
Figure 2
Relationship between the Fisher _P_-values and _Z_-scores for the muscle, liver and NF-κB reference sets. Based on the distribution of scores for the reference sets, a _Z_-score cutoff of 10 and a Fisher _P_-value cutoff of 0.01 were empirically selected as threshold levels to be used for testing. TFBSs that have functional relevance are labeled.
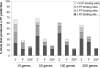
Figure 3
Percentage of trials that produced false positive (FP) predictions. Sets containing 15, 50, 100 and 200 randomly selected genes were generated and submitted to oPOSSUM (100 trials each). Each segment of the bar represents the percentage of trials where n TFBSs were over-represented by chance using the _Z_-score and Fisher _P_-value cutoffs. Symbols: Z = _Z_-score > 10; F = Fisher < 0.01; _Z_&_F_ = _Z_-score > 10 and Fisher < 0.01.
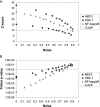
Figure 4
Noise tolerance. Increasing numbers of randomly selected genes were added to the muscle, liver and NF-κB reference sets to assess the effect of noise on (A) the _Z_-score and (B) Fisher exact probability statistical measures. The amount of noise is represented as the fraction of all genes in the set that were randomly selected. Average _Z_-scores and Fisher _P_-values for MEF2, HNF-1 and NF-κB over 100 trials for each noise level are shown to represent the muscle, liver and NF-κB reference sets, respectively. Suggested cutoffs for the _Z_-score and Fisher _P_-value are shown by the dotted grey lines.
Figure 5
The oPOSSUM result report for the identification of over-represented TFBSs in sets of co-expressed genes. (A) Results report showing the selected parameters, genes included and excluded in the analysis, and summary tables containing the Fisher exact probability scores and _Z_-scores for each TFBS (only the first five results are shown for each statistical test in this figure). (B) Pop-up window displaying genes that contain a particular TFBS (in this case, MEF2), as well as the site locations and scores.
Similar articles
- oPOSSUM: integrated tools for analysis of regulatory motif over-representation.
Ho Sui SJ, Fulton DL, Arenillas DJ, Kwon AT, Wasserman WW. Ho Sui SJ, et al. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W245-52. doi: 10.1093/nar/gkm427. Epub 2007 Jun 18. Nucleic Acids Res. 2007. PMID: 17576675 Free PMC article. - Genome-wide prediction of transcriptional regulatory elements of human promoters using gene expression and promoter analysis data.
Kim SY, Kim Y. Kim SY, et al. BMC Bioinformatics. 2006 Jul 4;7:330. doi: 10.1186/1471-2105-7-330. BMC Bioinformatics. 2006. PMID: 16817975 Free PMC article. - CONFAC: automated application of comparative genomic promoter analysis to DNA microarray datasets.
Karanam S, Moreno CS. Karanam S, et al. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W475-84. doi: 10.1093/nar/gkh353. Nucleic Acids Res. 2004. PMID: 15215433 Free PMC article. - CORE_TF: a user-friendly interface to identify evolutionary conserved transcription factor binding sites in sets of co-regulated genes.
Hestand MS, van Galen M, Villerius MP, van Ommen GJ, den Dunnen JT, 't Hoen PA. Hestand MS, et al. BMC Bioinformatics. 2008 Nov 26;9:495. doi: 10.1186/1471-2105-9-495. BMC Bioinformatics. 2008. PMID: 19036135 Free PMC article. - Cluster analysis and promoter modelling as bioinformatics tools for the identification of target genes from expression array data.
Werner T. Werner T. Pharmacogenomics. 2001 Feb;2(1):25-36. doi: 10.1517/14622416.2.1.25. Pharmacogenomics. 2001. PMID: 11258194 Review.
Cited by
- The dependence of expression of NF-κB-dependent genes: statistics and evolutionary conservation of control sequences in the promoter and in the 3' UTR.
Iwanaszko M, Brasier AR, Kimmel M. Iwanaszko M, et al. BMC Genomics. 2012 May 11;13:182. doi: 10.1186/1471-2164-13-182. BMC Genomics. 2012. PMID: 22577947 Free PMC article. - Uncovering cis-regulatory sequence requirements for context-specific transcription factor binding.
Yáñez-Cuna JO, Dinh HQ, Kvon EZ, Shlyueva D, Stark A. Yáñez-Cuna JO, et al. Genome Res. 2012 Oct;22(10):2018-30. doi: 10.1101/gr.132811.111. Epub 2012 Apr 25. Genome Res. 2012. PMID: 22534400 Free PMC article. - Nexus between epidermolysis bullosa and transcriptional regulation by thyroid hormone in epidermal keratinocytes.
Tomic-Canic M, Stojadinovic O, Lee B, Walsh R, Blumenberg M. Tomic-Canic M, et al. Clin Transl Sci. 2008 May;1(1):45-9. doi: 10.1111/j.1752-8062.2008.00015.x. Clin Transl Sci. 2008. PMID: 20443817 Free PMC article. - Molecular basis for hair loss in mice carrying a novel nonsense mutation (Hrrh-R ) in the hairless gene (Hr).
Liu Y, Sundberg JP, Das S, Carpenter D, Cain KT, Michaud EJ, Voy BH. Liu Y, et al. Vet Pathol. 2010 Jan;47(1):167-76. doi: 10.1177/0300985809352970. Vet Pathol. 2010. PMID: 20080498 Free PMC article. - Global analysis of estrogen receptor beta binding to breast cancer cell genome reveals an extensive interplay with estrogen receptor alpha for target gene regulation.
Grober OM, Mutarelli M, Giurato G, Ravo M, Cicatiello L, De Filippo MR, Ferraro L, Nassa G, Papa MF, Paris O, Tarallo R, Luo S, Schroth GP, Benes V, Weisz A. Grober OM, et al. BMC Genomics. 2011 Jan 14;12:36. doi: 10.1186/1471-2164-12-36. BMC Genomics. 2011. PMID: 21235772 Free PMC article.
References
- Stormo G.D. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. - PubMed
- Bulyk M.L., Gentalen E., Lockhart D.J., Church G.M. Quantifying DNA–protein interactions by double-stranded DNA arrays. Nat. Biotechnol. 1999;17:573–577. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources