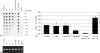A minor fraction of base J in kinetoplastid nuclear DNA is bound by the J-binding protein 1 - PubMed (original) (raw)
A minor fraction of base J in kinetoplastid nuclear DNA is bound by the J-binding protein 1
Cristiane Bentin Toaldo et al. Mol Biochem Parasitol. 2005 Sep.
No abstract available
Figures
Fig. 1
Determination of JBP1 levels in bloodstream-form T. brucei wild type HN221 (enES2) [18], JBP1 double knock out cell line JBP1dKO-4b1 [13], and JBP1 overexpressor (JBP1-neo), L. tarentolae and C. fasciculata using a semi-quantitative western blot. (A) Serial dilutions of sonicated total cell lysates and defined amounts of rJBP1 mixed in with carrier lysates (comparable to approximately 5 × 106 trypanosome cell equivalents/sample) were separated on an 8% SDS–polyacrylamide gel. After electrophoresis the proteins were blotted onto nitrocellulose and incubated with rabbit polyclonal antibodies directed against the respective JBP1 proteins. As JBP1 was hardly detectable in wild-type T. brucei, we made a cell line overexpressing JBP1. The JBP1 gene was amplified using construct pJBP-Phleo [13] and oligonucleotides (5′-GCAGTACTATGCGAAGGCAGGTGAAGAAGG and 5′-GCAGTACTCTATTTCTCCCGTCTGCGG) containing _Sca_I restriction sites. The JBP1 gene was introduced in the _Eco_RV site of the vector pURXN downstream of the ribosomal promoter and upstream of a NEOMYCIN gene [19]. This construct contains TUBULIN intergenic regions for correct RNA processing of both genes. The final construct pUR-JBP1-neo was transfected into JBP1dKO-4b1 bloodstream-form trypanosomes resulting in the cell line JBP1-neo. rJBP1 from each of the three species was expressed in E. coli and purified using a N-terminal His-tag [9]. For the expression of recombinant C. fasciculata JBP1 we replaced the wild-type gene with a mutant version of the gene encoding a protein in which the amino acids at positions 255, 256, 257 or 433 were substituted with alanines. C. fasciculata and L. tarentolae rJBP1 were purified as previously described [9]. T. brucei rJBP1 was expressed using BL 21 codon plus RIL E. coli grown at 37 °C to an OD600 of 0.6. JBP1 expression was induced for 2 h by the addition of 1 mM IPTG. Cells were collected by centrifugation and resuspended in lysis buffer (8 M urea; 1 M NaCl; 50 mM NaPO4 pH 7.2). The rJBP1 was purified using its His-tag [9] and further purified by gel electrophoresis and electro elution. Polyclonal antibodies against wild-type JBP1 from L. tarentolae and C. fasciculata were generated by cloning the full-length gene in the pET15b vector, producing N-terminally His tagged JBP1 [9]. Due to insufficient yield of full-length T. brucei recombinant protein, antiserum against T. brucei JBP1 was raised using a fragment of JBP1 encoding a polypeptide from amino acid 446 to amino acid 633. The fragment was cloned into the pMal-C vector (New England Biolabs) resulting in a maltose binding fusion protein. Rabbits were immunized with purified protein and after four boosts the sera were collected. A secondary HRP-labeled swine anti-rabbit antibody was used for detection in combination with ECL. JBP1 protein bands are indicated by arrowheads. Molecular weight markers (kDa) are shown on the right. (B) Quantitation of two independent experiments using independently prepared lysates. From the signal obtained with the rabbit JBP1 antibodies we estimate that T. brucei contains 0.40 ng JBP1 per 1 × 106 cells, L. tarentolae 0.19 ng JBP1 per 1 × 106 cells and C. fasciculata 0.16 ng JBP1 per 1 × 106 cells. Using the species specific JBP1 molecular weight (90–100 kDa) we calculate the number of JBP1 molecules per cell to be approximately 2600, 1200 and 1000 for T. brucei, L. tarentolae and C. fasciculate, respectively. Because the T. brucei JBP1 signal is very weak, we also determined the amount of JBP1 in a T. brucei JBP1-neo line, which overexpresses JBP1 4.5-fold (see text for explanation) and we calculated the amount of JBP1 in this strain to be approximately 11,700 JBP1 molecules per cell.
Fig. 2
Quantitation of J levels in the DNA of T. brucei, L. major, L. tarentolae and C. fasciculata using anti-J DNA immunoblots [5]. (A) Dot blots with two-fold dilution series of DNA were incubated with antiserum against J. Bound antibodies were detected by a second antibody conjugated to HRP and visualized by ECL. The DNAs used are from T. brucei (Tb) bloodstream form wild type HN221, L. major (Lm), L. tarentolae (Lt), C. fasciculata (Cf), T. brucei JBP1 knockout JBP1dKO-4b1, T. brucei JBP1 overexpressor JBP1-neo and T. brucei procyclic form (Tb PC), the last one included as a negative control. (B) Equal loading of the dot blots was confirmed by staining the DNA with ethidium bromide on an agarose gel. (C) T. brucei wild-type J level in the genome was set at 0.12% [4] and all the other species were plotted relative to T. brucei (0.11 ± 0.05% for L. major, 0.11 ± 0.04% for L. tarentolae, 0.05 ± 0.02% for C. fasciculata, 0.01 ± 0.01% for T. brucei JBP1dKO-4b1 and 0.22 ± 0.03% for T. brucei JBP1-neo; mean ± standard deviation based on at least four independent experiments).
Similar articles
- The modified base J is the target for a novel DNA-binding protein in kinetoplastid protozoans.
Cross M, Kieft R, Sabatini R, Wilm M, de Kort M, van der Marel GA, van Boom JH, van Leeuwen F, Borst P. Cross M, et al. EMBO J. 1999 Nov 15;18(22):6573-81. doi: 10.1093/emboj/18.22.6573. EMBO J. 1999. PMID: 10562569 Free PMC article. - Base J: discovery, biosynthesis, and possible functions.
Borst P, Sabatini R. Borst P, et al. Annu Rev Microbiol. 2008;62:235-51. doi: 10.1146/annurev.micro.62.081307.162750. Annu Rev Microbiol. 2008. PMID: 18729733 Review. - Two thymidine hydroxylases differentially regulate the formation of glucosylated DNA at regions flanking polymerase II polycistronic transcription units throughout the genome of Trypanosoma brucei.
Cliffe LJ, Siegel TN, Marshall M, Cross GA, Sabatini R. Cliffe LJ, et al. Nucleic Acids Res. 2010 Jul;38(12):3923-35. doi: 10.1093/nar/gkq146. Epub 2010 Mar 9. Nucleic Acids Res. 2010. PMID: 20215442 Free PMC article. - Telomeric localization of the modified DNA base J in the genome of the protozoan parasite Leishmania.
Genest PA, Ter Riet B, Cijsouw T, van Luenen HG, Borst P. Genest PA, et al. Nucleic Acids Res. 2007;35(7):2116-24. doi: 10.1093/nar/gkm050. Epub 2007 Feb 28. Nucleic Acids Res. 2007. PMID: 17329373 Free PMC article. - Protean permeases: Diverse roles for membrane transport proteins in kinetoplastid protozoa.
Landfear SM. Landfear SM. Mol Biochem Parasitol. 2019 Jan;227:39-46. doi: 10.1016/j.molbiopara.2018.12.006. Epub 2018 Dec 24. Mol Biochem Parasitol. 2019. PMID: 30590069 Free PMC article. Review.
Cited by
- Kinetic Studies on the 2-Oxoglutarate/Fe(II)-Dependent Nucleic Acid Modifying Enzymes from the AlkB and TET Families.
Peng Z, Ma J, Christov CZ, Karabencheva-Christova T, Lehnert N, Li D. Peng Z, et al. DNA (Basel). 2023 Jun;3(2):65-84. doi: 10.3390/dna3020005. Epub 2023 Mar 30. DNA (Basel). 2023. PMID: 38698914 Free PMC article. - A method for the efficient and selective identification of 5-hydroxymethyluracil in genomic DNA.
Bullard W, Kieft R, Sabatini R. Bullard W, et al. Biol Methods Protoc. 2017 Jan;2(1):bpw006. doi: 10.1093/biomethods/bpw006. Epub 2017 Jan 25. Biol Methods Protoc. 2017. PMID: 29276783 Free PMC article. - Base J glucosyltransferase does not regulate the sequence specificity of J synthesis in trypanosomatid telomeric DNA.
Bullard W, Cliffe L, Wang P, Wang Y, Sabatini R. Bullard W, et al. Mol Biochem Parasitol. 2015 Dec;204(2):77-80. doi: 10.1016/j.molbiopara.2016.01.005. Epub 2016 Jan 24. Mol Biochem Parasitol. 2015. PMID: 26815240 Free PMC article. - Defining the sequence requirements for the positioning of base J in DNA using SMRT sequencing.
Genest PA, Baugh L, Taipale A, Zhao W, Jan S, van Luenen HG, Korlach J, Clark T, Luong K, Boitano M, Turner S, Myler PJ, Borst P. Genest PA, et al. Nucleic Acids Res. 2015 Feb 27;43(4):2102-15. doi: 10.1093/nar/gkv095. Epub 2015 Feb 6. Nucleic Acids Res. 2015. PMID: 25662217 Free PMC article. - Identification of the glucosyltransferase that converts hydroxymethyluracil to base J in the trypanosomatid genome.
Bullard W, Lopes da Rosa-Spiegler J, Liu S, Wang Y, Sabatini R. Bullard W, et al. J Biol Chem. 2014 Jul 18;289(29):20273-82. doi: 10.1074/jbc.M114.579821. Epub 2014 Jun 2. J Biol Chem. 2014. PMID: 24891501 Free PMC article.
References
- Gommers-Ampt JH, Van Leeuwen F, De Beer AL, Vliegenthart JF, Dizdaroglu M, Kowalak JA, et al. β-D-Glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75:1129–36. - PubMed
- Van Leeuwen F, Kieft R, Cross M, Borst P. Tandemly repeated DNA is a target for the partial replacement of thymine by β-D-glucosyl-hydoxymethyluracil in Trypanosoma brucei. Mol Biochem Parasitol. 2000;109:133–45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials