The primary mechanism of the IL-10-regulated antiinflammatory response is to selectively inhibit transcription - PubMed (original) (raw)
Comparative Study
. 2005 Jun 14;102(24):8686-91.
doi: 10.1073/pnas.0500419102. Epub 2005 Jun 3.
Affiliations
- PMID: 15937121
- PMCID: PMC1150817
- DOI: 10.1073/pnas.0500419102
Comparative Study
The primary mechanism of the IL-10-regulated antiinflammatory response is to selectively inhibit transcription
Peter J Murray. Proc Natl Acad Sci U S A. 2005.
Abstract
The antiinflammatory cytokine IL-10 inhibits the production of multiple, diverse inflammatory mediators from activated macrophages and dendritic cells, a process requiring STAT3 activation. However, the mechanisms involved in the broad inhibitory effects of IL-10 are controversial. I eliminated the contribution of the major confounding variable to understanding the antiinflammatory response, the 3' UTR region of inflammatory mediator genes, through knock-in mutation and analysis of the effects of IL-10 on transcription rate of inflammatory genes. IL-10 activates STAT3 to act indirectly by selectively inhibiting gene transcription independent of general effects on NF-kappaB or posttranscriptional mRNA processing through a process that reduces the overall transcriptional rate of specific genes.
Figures
Fig. 1.
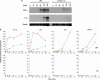
IL-10 inhibits the rate of transcription at LPS-induced inflammatory genes. (a) Total RNA was isolated from stimulated macrophages and subjected to denaturing agarose gel electrophoresis to assess RNA quality followed by Northern blotting using probes specific for IL-10 targets whose expression levels have been previously shown to decrease after costimulation with LPS and IL-10. Shown is the same Northern blot probed with cDNA probes for TNF-α or IL-1α. Note that IL-10 strongly inhibits total mRNA levels. (b) PT RT-PCR assays were performed for the target genes shown by using normalized DNase-treated RNA samples from _IL-10_-/- BMDMs stimulated with 10 ng/ml IL-10 (black triangles), 100 ng/ml LPS (red triangles), or LPS and IL-10 (green triangles) for 0, 30, 60, 120, and 240 min. RNA was reverse-transcribed in the presence (Upper) or absence (Lower) of reverse transcriptase (RT) and subjected to SYBR green real-time PT RT-PCR. Results are representative of three independent experiments.
Fig. 2.
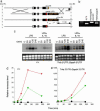
Analysis of the effects of IL-10 on IκB and IKK activity. (a) PT RT-PCR analysis of the effects of IL-10 on LPS-mediated induction of IκBα transcription. _IL-10_-/- BMDMs stimulated with 10 ng/ml IL-10 (black circles), 100 ng/ml LPS (green triangles) or LPS and IL-10 (red triangles) for 0, 30, 60, 120, and 240 min. RNA was reverse-transcribed and subjected to SYBR green real-time PT RT-PCR. (b) Immunoblotting analysis of IκBα and IκBβ levels in primary macrophages after stimulation with LPS and IL-10. Note that IκB proteins levels drop after proteosome-mediated destruction of each protein followed by resynthesis. (c) (Left and Center) IKK activity assays showing that IL-10 does not affect the ability of immunoprecipitated IKK to phosphorylate a GST-IκBα 1-55 purified recombinant fusion protein. (Right) The same assay using a mutant form of the substrate.
Fig. 3.
Construction and analysis of mice in which the Tnfa locus has been modified to replace the endogenous 3′ UTR with the Gapdh 3′ UTR. (a) Schematic diagram of the modified locus after targeting and subsequent cre-lox-mediated elimination of the pgk-neo cassette. (b) PCR analysis of mice carrying the wild-type or _Tnfa 3_′ UTR::_Gapdh 3_′ UTR alleles. (c) Northern blotting analysis using a TNF-α cDNA probe of macrophages from wild-type or _Tnfa 3_′ UTR::_Gapdh 3_′ UTR mice treated with LPS or LPS and IL-10 under conditions described in the legend to Fig. 1. Note that the modification of the Tnfa locus causes two transcripts to be made. Equivalent total RNA levels are shown by ethidium bromide staining. (d and e) Representative conventional real-time RT-PCR analysis from RNA samples isolated from stimulated BMDMs from wild type for _Tnfa 3_′ UTR::_Gapdh 3_′ UTR mice. BMDMs were stimulated with IL-10 (black circles), LPS (red triangles), or LPS and IL-10 (green triangles) as described in the legend to Fig. 1. Note the scale difference between the two genotypes reflected the higher levels of the stabilized TNF-α mRNA in BMDMs from _Tnfa 3_′ UTR::_Gapdh 3_′ UTR mice.
Fig. 4.
The IL-10-mediated antiinflammatory effect requires new protein synthesis. (a and b) PT RT-PCR analysis of IL-10 targets in the absence (a) or presence (b) of 10 μg/ml cycloheximide (CHX). Total RNA was isolated from _IL-10_-/- macrophages stimulated as described in Fig. 1 (LPS, green triangles; LPS and IL-10, red triangles) and subjected to PT RT-PCR. Note that induction of IL-6 by LPS requires new protein synthesis and that the requirement for new protein synthesis by IL-10 to block this gene cannot be determined. Data are representative of two independent experiments performed under identical conditions. (c) Northern blotting analysis of the effects of cycloheximide on IL-10-mediated inhibition of TNF-α mRNA in _Tnfa 3_′ UTR::_Gapdh 3_′ UTR BMDMs. Data are representative of three independent experiments. (d and e) Quantitative analysis by RT-PCR of the effects of IL-10 on _Tnfa 3_′ UTR::_Gapdh 3_′ UTR mRNA levels. Shown is conventional RT-PCR analysis for TNF-α using a FAM-labeled probe. BMDMs were stimulated with LPS (green triangles) or LPS and IL-10 (red triangles) as described in the legend to Fig. 1. Data were normalized by using β-actin as a standard. Data are representative of three independent experiments.
Similar articles
- An RNA-binding protein alphaCP-1 is involved in the STAT3-mediated suppression of NF-kappaB transcriptional activity.
Nishinakamura H, Minoda Y, Saeki K, Koga K, Takaesu G, Onodera M, Yoshimura A, Kobayashi T. Nishinakamura H, et al. Int Immunol. 2007 May;19(5):609-19. doi: 10.1093/intimm/dxm026. Epub 2007 Mar 22. Int Immunol. 2007. PMID: 17383969 - Inhibition of IL-10-induced STAT3 activation by 15-deoxy-Delta12,14-prostaglandin J2.
Ji JD, Kim HJ, Rho YH, Choi SJ, Lee YH, Cheon HJ, Sohn J, Song GG. Ji JD, et al. Rheumatology (Oxford). 2005 Aug;44(8):983-8. doi: 10.1093/rheumatology/keh657. Epub 2005 Apr 19. Rheumatology (Oxford). 2005. PMID: 15840591 - Inflammation and necrosis promote tumour growth.
Vakkila J, Lotze MT. Vakkila J, et al. Nat Rev Immunol. 2004 Aug;4(8):641-8. doi: 10.1038/nri1415. Nat Rev Immunol. 2004. PMID: 15286730 Review. No abstract available. - The IL-10/STAT3-mediated anti-inflammatory response: recent developments and future challenges.
Hutchins AP, Diez D, Miranda-Saavedra D. Hutchins AP, et al. Brief Funct Genomics. 2013 Nov;12(6):489-98. doi: 10.1093/bfgp/elt028. Epub 2013 Aug 12. Brief Funct Genomics. 2013. PMID: 23943603 Free PMC article. Review.
Cited by
- Genomic analysis of LPS-stimulated myeloid cells identifies a common pro-inflammatory response but divergent IL-10 anti-inflammatory responses.
Hutchins AP, Takahashi Y, Miranda-Saavedra D. Hutchins AP, et al. Sci Rep. 2015 Mar 13;5:9100. doi: 10.1038/srep09100. Sci Rep. 2015. PMID: 25765318 Free PMC article. - Acute infection of mice with Clostridium difficile leads to eIF2α phosphorylation and pro-survival signalling as part of the mucosal inflammatory response.
Sadighi Akha AA, Theriot CM, Erb-Downward JR, McDermott AJ, Falkowski NR, Tyra HM, Rutkowski DT, Young VB, Huffnagle GB. Sadighi Akha AA, et al. Immunology. 2013 Sep;140(1):111-22. doi: 10.1111/imm.12122. Immunology. 2013. PMID: 23668260 Free PMC article. - Interleukin-10 and Small Molecule SHIP1 Allosteric Regulators Trigger Anti-inflammatory Effects through SHIP1/STAT3 Complexes.
Chamberlain TC, Cheung ST, Yoon JSJ, Ming-Lum A, Gardill BR, Shakibakho S, Dzananovic E, Ban F, Samiea A, Jawanda K, Priatel J, Krystal G, Ong CJ, Cherkasov A, Andersen RJ, McKenna SA, Van Petegem F, Mui AL. Chamberlain TC, et al. iScience. 2020 Aug 21;23(8):101433. doi: 10.1016/j.isci.2020.101433. Epub 2020 Aug 2. iScience. 2020. PMID: 32823063 Free PMC article. - 9-cis retinoic acid inhibits inflammatory responses of adherent monocytes and increases their ability to induce classical monocyte migration.
Kolseth IB, Agren J, Sundvold-Gjerstad V, Lyngstadaas SP, Wang JE, Dahle MK. Kolseth IB, et al. J Innate Immun. 2012;4(2):176-86. doi: 10.1159/000332375. Epub 2011 Dec 29. J Innate Immun. 2012. PMID: 22213773 Free PMC article. - Single-cell analysis reveals TLR-induced macrophage heterogeneity and quorum sensing dictate population wide anti-inflammatory feedback in response to LPS.
Tiemeijer BM, Heester S, Sturtewagen AYW, Smits AIPM, Tel J. Tiemeijer BM, et al. Front Immunol. 2023 Feb 24;14:1135223. doi: 10.3389/fimmu.2023.1135223. eCollection 2023. Front Immunol. 2023. PMID: 36911668 Free PMC article.
References
- Bogdan, C., Paik, J., Vodovotz, Y. & Nathan, C. (1992) J. Biol. Chem. 267, 23301-23308. - PubMed
- Fiorentino, D. F., Zlotnik, A., Vieira, P., Mosmann, T. R., Howard, M., Moore, K. W. & O'Garra, A. (1991) J. Immunol. 146, 3444-3451. - PubMed
- Donnelly, R. P., Dickensheets, H. & Finbloom, D. S. (1999) J. Interferon Cytokine Res. 19, 563-573. - PubMed
- Moore, K. W., de Waal Malefyt, R., Coffman, R. L. & O'Garra, A. (2001) Annu. Rev. Immunol. 19, 683-765. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous