S-nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities - PubMed (original) (raw)
Comparative Study
. 2005 Jun 14;102(24):8525-30.
doi: 10.1073/pnas.0407294102. Epub 2005 Jun 3.
Affiliations
- PMID: 15937123
- PMCID: PMC1150803
- DOI: 10.1073/pnas.0407294102
Comparative Study
S-nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities
Antonio Martínez-Ruiz et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Nitric oxide is implicated in a variety of signaling pathways in different systems, notably in endothelial cells. Some of its effects can be exerted through covalent modifications of proteins and, among these modifications, increasing attention is being paid to S-nitrosylation as a signaling mechanism. In this work, we show by a variety of methods (ozone chemiluminescence, biotin switch, and mass spectrometry) that the molecular chaperone Hsp90 is a target of S-nitrosylation and identify a susceptible cysteine residue in the region of the C-terminal domain that interacts with endothelial nitric oxide synthase (eNOS). We also show that the modification occurs in endothelial cells when they are treated with S-nitroso-l-cysteine and when they are exposed to eNOS activators. Hsp90 ATPase activity and its positive effect on eNOS activity are both inhibited by S-nitrosylation. Together, these data suggest that S-nitrosylation may functionally regulate the general activities of Hsp90 and provide a feedback mechanism for limiting eNOS activation.
Figures
Fig. 1.
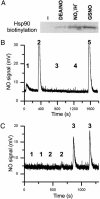
Detection of S-nitrosylation by the biotin switch technique and by specific reduction and ozone chemiluminescence. (A) Recombinant human Hsp90α (1.3 μM) was left untreated (-) or treated with 1 mM diethylamine-NONOate (DEA/NO), 2.7 μM NaNO2 in 0.5 M HCl (NO2-/H+), or 1 mM GSNO. Hsp90α preparations were then subjected to the biotin switch assay, and biotinylated protein was detected by Western blot with streptavidin. The image shown is representative of three independent experiments. (B and C) Recombinant human Hsp90α was reduced with Cu+/cysteine, and NO release detected in a nitric oxide analyzer apparatus. Figures shown are representative of at least three independent experiments. (B) Protein (5.3 μM) or buffer was treated with 1 mM GSNO, and protein and low molecular-mass (LMM) fractions were separated in a Sephadex G-25 column before analysis. 1: Hsp90 plus GSNO, protein fraction; 2: Hsp90 plus GSNO, LMM fraction; 3: Hsp90, protein fraction; 4: buffer plus GSNO, protein fraction; 5: buffer plus GSNO, LMM fraction. (C) Protein (0.8 μM) was treated with 1.6 μM NaNO2 in 0.5 M HCl or HCl only for 20 min and adjusted to pH 7.4 with NaOH and PBS before analysis. 1: Buffer plus NaNO2; 2: protein without NaNO2; 3: protein plus NaNO2.
Fig. 2.
Detection and identification of S-nitrosylation by mass spectrometry. (A) Recombinant human Hsp90α was either treated with NaNO2 (1:2 molar ratio) in 0.5 M HCl and adjusted to pH 7.4 with NaOH and PBS or with 1 mM GSNO in PBS. Samples were then trypsin digested and analyzed by LC-MS by using a linear ion trap detector, as described in Materials and Methods. Shown is the fragmentation spectrum of the S-nitrosylated peptide corresponding to residues 591–611 of Hsp90α, together with the assignation of the fragmentation series to the sequence of the modified peptide. *, tentative S-nitrosylated Cys. (B) Schematic of Hsp90α domains (in boxes) and the localization of the cysteine residues (dots) and the S-nitrosylated peptide detected by MS. Functions assigned to the domains are written above, and regions described to interact with Akt and eNOS are shown below. Numbers correspond to the human Hsp90α sequence and, in the case of the domains, they denote the boundaries of the crystallographic structures. (C) Ribbon representation of the C-domain structure model of human Hsp90α (residues 568 to 693 of
swiss
-
prot
entry P07900), based on the structure of the C-terminal domain of E. coli htpG (PDB ID code 1fs8D). The side chains of cysteines 596 and 597, arginine 590, and glutamic acid 634 are highlighted.
Fig. 3.
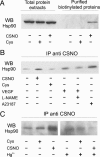
Detection of S-nitrosylated Hsp90 in endothelial cells. (A) EA.hy926 cells were untreated (-) or exposed (+) to 1 mM cysteine (Cys) or S-nitrosocysteine (CSNO) for 15 min. Protein extracts were subjected to the biotin switch technique, and the biotinylated proteins were purified with avidin-agarose, followed by elution with 2-mercaptoethanol. Hsp90 was detected in total protein extracts and in eluates by SDS/PAGE and immunoblot. (B) EA.hy926 cells were untreated, treated with Cys or CSNO (1 mM), or stimulated with VEGF (50 ng/ml) with or without prior incubation with L-NAME (0.5 mM, 1 h) or calcium ionophore A23187 (10 mM) for 15 min. Protein extracts were immunoprecipitated with anti S-nitrosocysteine and Hsp90 identified by immunoblot. (C) Extracts from EA.hy926 cells untreated or exposed to Cys or CSNO (1 mM) were immunoprecipitated with anti-S-nitrosocysteine with or without prior specific breakdown of the S-NO bond by Hg2+ and Hsp90 identified by immunoblot. Figures shown are representative of at least two independent experiments.
Fig. 4.
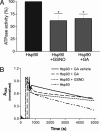
Hsp90 ATPase activity is inhibited by S-nitrosylation. Hsp90 was 1 μM throughout. (A) Activities are shown for Hsp90 alone, after S-nitrosylation with 1 mM GSNO (n = 7), or incubation with the Hsp90-specific inhibitor, geldanamycin (GA; 2 μM, n = 4). ATPase activity is expressed as a relative slope to that of Hsp90 alone. Values are means ± SEM; *, P < 0.05 with respect to Hsp90 alone. (B) A representative experiment showing experimental progress curves corresponding to NADH consumption after addition of Hsp90 alone (in Tris buffer), Hsp90 plus a GA vehicle (DMSO), Hsp90 preincubated with GSNO, or Hsp90 plus GA.
Fig. 5.
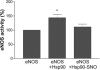
eNOS activation by Hsp90 is inhibited by S-nitrosylation. Purified eNOS (1 μM) was incubated with or without native or S-nitrosylated Hsp90 (1 μM), and eNOS activity was determined from the conversion of 3H-
l
-arginine to 3H-
l
-citrulline. Data are means ± SEM, n = 7 experiments in duplicate; *, P < 0.01 with respect to eNOS alone.
Similar articles
- Hsp90 S-nitrosylation at Cys521, as a conformational switch, modulates cycling of Hsp90-AHA1-CDC37 chaperone machine to aggravate atherosclerosis.
Zhao S, Tang X, Miao Z, Chen Y, Cao J, Song T, You D, Zhong Y, Lin Z, Wang D, Shi Z, Tang X, Wang D, Chen S, Wang L, Gu A, Chen F, Xie L, Huang Z, Wang H, Ji Y. Zhao S, et al. Redox Biol. 2022 Jun;52:102290. doi: 10.1016/j.redox.2022.102290. Epub 2022 Mar 17. Redox Biol. 2022. PMID: 35334246 Free PMC article. - Shear flow increases S-nitrosylation of proteins in endothelial cells.
Huang B, Chen SC, Wang DL. Huang B, et al. Cardiovasc Res. 2009 Aug 1;83(3):536-46. doi: 10.1093/cvr/cvp154. Epub 2009 May 15. Cardiovasc Res. 2009. PMID: 19447776 - Antioxidant effects of statins via S-nitrosylation and activation of thioredoxin in endothelial cells: a novel vasculoprotective function of statins.
Haendeler J, Hoffmann J, Zeiher AM, Dimmeler S. Haendeler J, et al. Circulation. 2004 Aug 17;110(7):856-61. doi: 10.1161/01.CIR.0000138743.09012.93. Epub 2004 Aug 2. Circulation. 2004. PMID: 15289372 - Protein s-nitrosylation measurement.
Qin Y, Dey A, Daaka Y. Qin Y, et al. Methods Enzymol. 2013;522:409-25. doi: 10.1016/B978-0-12-407865-9.00019-4. Methods Enzymol. 2013. PMID: 23374195 Review. - Post-translational control of endothelial nitric oxide synthase: why isn't calcium/calmodulin enough?
Fulton D, Gratton JP, Sessa WC. Fulton D, et al. J Pharmacol Exp Ther. 2001 Dec;299(3):818-24. J Pharmacol Exp Ther. 2001. PMID: 11714864 Review.
Cited by
- Identification of Protein Targets of _S_-Nitroso-Coenzyme A-Mediated _S_-Nitrosation Using Chemoproteomics.
Falco JA, Wynia-Smith SL, McCoy J, Smith BC, Weerapana E. Falco JA, et al. ACS Chem Biol. 2024 Jan 19;19(1):193-207. doi: 10.1021/acschembio.3c00654. Epub 2023 Dec 30. ACS Chem Biol. 2024. PMID: 38159293 - HSP90 at the hub of protein homeostasis: emerging mechanistic insights.
Taipale M, Jarosz DF, Lindquist S. Taipale M, et al. Nat Rev Mol Cell Biol. 2010 Jul;11(7):515-28. doi: 10.1038/nrm2918. Epub 2010 Jun 9. Nat Rev Mol Cell Biol. 2010. PMID: 20531426 Review. - Desensitization of soluble guanylyl cyclase, the NO receptor, by S-nitrosylation.
Sayed N, Baskaran P, Ma X, van den Akker F, Beuve A. Sayed N, et al. Proc Natl Acad Sci U S A. 2007 Jul 24;104(30):12312-7. doi: 10.1073/pnas.0703944104. Epub 2007 Jul 16. Proc Natl Acad Sci U S A. 2007. PMID: 17636120 Free PMC article. - Computational Structural Biology of _S_-nitrosylation of Cancer Targets.
Bignon E, Allega MF, Lucchetta M, Tiberti M, Papaleo E. Bignon E, et al. Front Oncol. 2018 Aug 14;8:272. doi: 10.3389/fonc.2018.00272. eCollection 2018. Front Oncol. 2018. PMID: 30155439 Free PMC article. Review. - Regulation by Different Types of Chaperones of Amyloid Transformation of Proteins Involved in the Development of Neurodegenerative Diseases.
Muronetz VI, Kudryavtseva SS, Leisi EV, Kurochkina LP, Barinova KV, Schmalhausen EV. Muronetz VI, et al. Int J Mol Sci. 2022 Mar 2;23(5):2747. doi: 10.3390/ijms23052747. Int J Mol Sci. 2022. PMID: 35269889 Free PMC article. Review.
References
- Stamler, J. S., Lamas, S. & Fang, F. C. (2001) Cell 106, 675-683. - PubMed
- Martínez-Ruiz, A. & Lamas, S. (2004) Cardiovasc. Res. 62, 43-52. - PubMed
- Fleming, I. & Busse, R. (2003) Am. J. Physiol. 284, R1-R12. - PubMed
- Sessa, W. C. (2004) J. Cell Sci. 117, 2427-2429. - PubMed
- Martínez-Ruiz, A. & Lamas, S. (2004) Arch. Biochem. Biophys. 423, 192-199. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases