Genome amplification of single sperm using multiple displacement amplification - PubMed (original) (raw)
Genome amplification of single sperm using multiple displacement amplification
Zhengwen Jiang et al. Nucleic Acids Res. 2005.
Abstract
Sperm typing is an effective way to study recombination rate on a fine scale in regions of interest. There are two strategies for the amplification of single meiotic recombinants: repulsion-phase allele-specific PCR and whole genome amplification (WGA). The former can selectively amplify single recombinant molecules from a batch of sperm but is not scalable for high-throughput operation. Currently, primer extension pre-amplification is the only method used in WGA of single sperm, whereas it has limited capacity to produce high-coverage products enough for the analysis of local recombination rate in multiple large regions. Here, we applied for the first time a recently developed WGA method, multiple displacement amplification (MDA), to amplify single sperm DNA, and demonstrated its great potential for producing high-yield and high-coverage products. In a 50 mul reaction, 76 or 93% of loci can be amplified at least 2500- or 250-fold, respectively, from single sperm DNA, and second-round MDA can further offer >200-fold amplification. The MDA products are usable for a variety of genetic applications, including sequencing and microsatellite marker and single nucleotide polymorphism (SNP) analysis. The use of MDA in single sperm amplification may open a new era for studies on local recombination rates.
Figures
Figure 1
PCR amplification of the three fragments of the TOP1, P53 and CYP1A2 genes in MDA products of 16 sperm aliquots treated using two different lysis methods. The fragments are 1080 bp in length for TOP1, 643 bp for P53 and 550 bp for CYP1A2. Eight aliquots from the dilution of 3 cells/3 μl were lysed on ice and the other eight were lysed at 65°C.
Figure 2
PCR amplification of the three fragments of the TOP1, P53 and CYP1A2 genes in MDA products of S01–S16 samples. The fragments are 1080 bp in length for TOP1, 643 bp for P53 and 550 bp for CYP1A2. S01–S16 indicate MDA products from 16 sperm aliquots. S16MDA2 is the second-round MDA product using 1 μl of 1/10C0 S16 MDA product as resource DNA. BLANK here is the MDA product of the negative control, in which 3 μl PBS buffer was added instead of sperm aliquot.
Figure 3
PCR amplification of 12 fragments using 1 μl of 1/5C0 or 1/50C0 MDA products as templates. The amplification result is indicated with a black square for successful amplification in 1/50C0, a gray square for successful amplification in 1/5C0 and a framed F for no amplification in both.
Figure 4
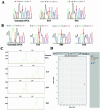
MDA products were useful for a variety of genetic applications, including sequencing and SNP and STR analysis. (A) A mutation at 45803241 of NT_010194.16 is marked by red arrows in the S14 MDA product. (B) Sequencing traces for rs1048943 (marked by red arrows) and its flanking 10 nt in genomic DNA, S14, S06 and S16 MDA products. S06 has two alleles, although the C allele was much preferred for amplification. (C) The electropherogram for the D10S547 locus in genomic DNA, S14, S04 and S05 samples displayed by GeneMapper V3.0. Their genotypes were displayed in Table 5. S14 was observed to have two alleles (244 and 254), although allele 254 showed to be preferentially amplified. The orange peak is a 250 bp marker. (D) The interface for the identification of genotypes at rs1535721 locus by Taqman assay using the ABI7900. The green one is for genomic DNA sample, blue for S05, S13 and S15, and red for S06, S08, S14, S16 and S16MDA2. × is for S04, for which the amplification failed.
Similar articles
- Multiple displacement amplification to create a long-lasting source of DNA for genetic studies.
Lovmar L, Syvänen AC. Lovmar L, et al. Hum Mutat. 2006 Jul;27(7):603-14. doi: 10.1002/humu.20341. Hum Mutat. 2006. PMID: 16786504 Review. - Single-nucleotide-polymorphism genotyping for whole-genome-amplified samples using automated fluorescence correlation spectroscopy.
Bannai M, Higuchi K, Akesaka T, Furukawa M, Yamaoka M, Sato K, Tokunaga K. Bannai M, et al. Anal Biochem. 2004 Apr 15;327(2):215-21. doi: 10.1016/j.ab.2004.01.012. Anal Biochem. 2004. PMID: 15051538 - Genome-wide single-nucleotide polymorphism arrays demonstrate high fidelity of multiple displacement-based whole-genome amplification.
Tzvetkov MV, Becker C, Kulle B, Nürnberg P, Brockmöller J, Wojnowski L. Tzvetkov MV, et al. Electrophoresis. 2005 Feb;26(3):710-5. doi: 10.1002/elps.200410121. Electrophoresis. 2005. PMID: 15690424 - Improved multiple displacement amplification with phi29 DNA polymerase for genotyping of single human cells.
Kumar G, Garnova E, Reagin M, Vidali A. Kumar G, et al. Biotechniques. 2008 Jun;44(7):879-90. doi: 10.2144/000112755. Biotechniques. 2008. PMID: 18533898 - Whole genome amplification from a single cell: a new era for preimplantation genetic diagnosis.
Coskun S, Alsmadi O. Coskun S, et al. Prenat Diagn. 2007 Apr;27(4):297-302. doi: 10.1002/pd.1667. Prenat Diagn. 2007. PMID: 17278176 Review.
Cited by
- Variants in the HEPSIN gene are associated with prostate cancer in men of European origin.
Pal P, Xi H, Kaushal R, Sun G, Jin CH, Jin L, Suarez BK, Catalona WJ, Deka R. Pal P, et al. Hum Genet. 2006 Sep;120(2):187-92. doi: 10.1007/s00439-006-0204-3. Epub 2006 Jun 17. Hum Genet. 2006. PMID: 16783571 - Use of multiple displacement amplification in the investigation of human papillomavirus physical status.
Evans MF, Adamson CS, von Walstrom GM, Cooper K. Evans MF, et al. J Clin Pathol. 2007 Oct;60(10):1135-9. doi: 10.1136/jcp.2006.041392. Epub 2006 Dec 20. J Clin Pathol. 2007. PMID: 17182658 Free PMC article. - Amplification of multiple genomic loci from single cells isolated by laser micro-dissection of tissues.
Frumkin D, Wasserstrom A, Itzkovitz S, Harmelin A, Rechavi G, Shapiro E. Frumkin D, et al. BMC Biotechnol. 2008 Feb 20;8:17. doi: 10.1186/1472-6750-8-17. BMC Biotechnol. 2008. PMID: 18284708 Free PMC article. - Evaluation of single-cell genomics to address evolutionary questions using three SAGs of the choanoflagellate Monosiga brevicollis.
López-Escardó D, Grau-Bové X, Guillaumet-Adkins A, Gut M, Sieracki ME, Ruiz-Trillo I. López-Escardó D, et al. Sci Rep. 2017 Sep 8;7(1):11025. doi: 10.1038/s41598-017-11466-9. Sci Rep. 2017. PMID: 28887541 Free PMC article. - Characterization of whole genome amplified (WGA) DNA for use in genotyping assay development.
Han T, Chang CW, Kwekel JC, Chen Y, Ge Y, Martinez-Murillo F, Roscoe D, Težak Z, Philip R, Bijwaard K, Fuscoe JC. Han T, et al. BMC Genomics. 2012 Jun 1;13:217. doi: 10.1186/1471-2164-13-217. BMC Genomics. 2012. PMID: 22655855 Free PMC article.
References
- Goldstein D.B. Islands of linkage disequilibrium. Nature Genet. 2001;29:109–111. - PubMed
- Daly M.J., Rioux J.D., Schaffner S.F., Hudson T.J., Lander E.S. High-resolution haplotype structure in the human genome. Nature Genet. 2001;29:229–232.. - PubMed
- Jeffreys A.J., Kauppi L., Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nature Genet. 2001;29:217–222. - PubMed
- Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. - PubMed
- Nejentsev S., Godfrey L., Snook H., Rance H., Nutland S., Walker N.M., Lam A.C., Guja C., Ionescu-Tirgoviste C., Undlien D.E., et al. Comparative high-resolution analysis of linkage disequilibrium and tag single nucleotide polymorphisms between populations in the vitamin D receptor gene. Hum. Mol. Genet. 2004;13:1633–1639. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources