ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif - PubMed (original) (raw)
ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif
Joëlle Bigay et al. EMBO J. 2005.
Abstract
ArfGAP1 promotes GTP hydrolysis in Arf1, a small G protein that interacts with lipid membranes and drives the assembly of the COPI coat in a GTP-dependent manner. The activity of ArfGAP1 increases with membrane curvature, suggesting a negative feedback loop in which COPI-induced membrane deformation determines the timing and location of GTP hydrolysis within a coated bud. Here we show that a central sequence of about 40 amino acids in ArfGAP1 acts as a lipid-packing sensor. This ALPS motif (ArfGAP1 Lipid Packing Sensor) is also found in the yeast homologue Gcs1p and is necessary for coupling ArfGAP1 activity with membrane curvature. The ALPS motif binds avidly to small liposomes and shows the same hypersensitivity on liposome radius as full-length ArfGAP1. Site-directed mutagenesis, limited proteolysis and circular dichroism experiments suggest that the ALPS motif, which is unstructured in solution, inserts bulky hydrophobic residues between loosely packed lipids and forms an amphipathic helix on highly curved membranes. This helix differs from classical amphipathic helices by the abundance of serine and threonine residues on its polar face.
Figures
Figure 1
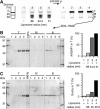
Gcs1p is sensitive to membrane curvature. (A) Sequence alignment of the central region of ArfGAP1 orthologues, including yeast Gcs1p. The abbreviations used are as follows: H.s. Homo sapiens, R.n. Rattus norvegicus, X.l. Xenopus laevis, D.m. Drosophila melanogaster, C.e. Caenorhabditis elegans, S.c. Saccharomyces cerevisiae. The Uniprot accession numbers are indicated. Black arrows indicate point mutations introduced in rat ArfGAP1 and the white arrow indicates a cleavage site by chymotrypsin. Conserved residues are highlighted with the following colour code: yellow, hydrophobic; purple, serine and threonine; blue, basic; red, acidic; grey, other residues. (B) Time course of GTP hydrolysis in Arf1 (0.5 μM) on Golgi-mix (C18:1–C18) liposomes of defined size upon the addition of Gcs1p (50 nM). The reaction was followed by tryptophan fluorescence. The hydrodynamic radius of the liposomes was determined by dynamic light scattering (DLS) and is indicated. (C) Rate (1/_t_1/2) of GTP hydrolysis in Arf1 as a function of the liposome radius in the presence of 50 nM Gcs1p (black circles) or 50 nM ArfGAP1 (white circles). Golgi-mix liposomes were made from natural lipids or enriched in C18:1–C18:1 lipids (see Materials and methods).
Figure 2
The binding ArfGAP1 and Gcs1p to liposomes increases with membrane curvature. (A) Flotation assay. ArfGAP1 (0.75 μM) was incubated with Golgi-mix liposomes (0.75 mM lipids) of decreasing size (tubes 2–4) or with no liposome (tube 1). The suspension was adjusted to 30% w/v sucrose and overlaid with two cushions of decreasing sucrose density. The NBD fluorescence of the liposomes was visualized in the tubes before and after centrifugation using a fluorescence imaging system. NBD fluorescence appears as black. (B) After centrifugation, the top (T), middle (M) and bottom (B) fractions were collected and analysed by SDS–PAGE. Proteins were stained with sypro-orange. The 100% lane was used to determine the ratio of ArfGAP1 present in the top fraction (right panel). (C) Same analysis with Gcs1p using C18:1–C18:1 Golgi-mix liposomes.
Figure 3
The central region of ArfGAP1 is necessary and sufficient to sense membrane curvature. (A) Diagram of the various constructs. (B) SDS–PAGE of the purified proteins visualized with sypro-orange. Note the presence of a degradation product in the GST constructs (white arrow). This form probably corresponds to a GST-192–214 fusion form as assessed by HPLC and mass spectrometry analysis (see Materials and methods). (C) Size-exclusion chromatography of the constructs with a polyhistidine tag on a superose-12 column. The straight line represents the best fit for the elution of molecular weight standards. The grey square (1–257AAA) represents the elution peak of [1–257]ArfGAP1 carrying the triple L207A–W211A–F214A mutation. (D) Avidity of the various constructs for Golgi-mix liposomes of decreasing size. For all constructs, flotation experiments similar to that described in Figure 2 were conducted except that the protein concentration was 1 μM and the lipid concentration 0.5 mM. The error bars show the variation observed between two independent experiments using different batches of liposomes. A detail view of the SDS–PAGE is shown for some GST constructs. (E) GAP assay using Arf-GTP bound to Golgi-mix liposomes of defined size. ArfGAP1[1–148] and ArfGAP1[1–196] were used at 100 nM. ArfGAP1[1–257] was used at 50 nM.
Figure 4
The interaction between the central region of ArfGAP1 and curved lipid membranes is exclusively hydrophobic. (A) Avidity of [137–257]ArfGAP1 for neutral or anionic liposomes of defined size as assessed by flotation experiments. The liposomes contained 100% DOPC (white circles), 100% POPC (white squares), 70% DOPC and 30% DOPS (black circles) or 70% POPC and 30% POPS (black squares). (B) GAP assay using Arf1-GTP bound to Golgi-mix liposomes extruded through 0.2 μm (grey traces) or 0.03 μm (black traces) polycarbonate filters. The hydrodynamic radius of the liposomes was 92 and 33 nm, respectively. GTP hydrolysis was initiated by the addition of 50 nM [1–257]ArfGAP1 carrying the indicated point mutations and was followed by tryptophan fluorescence. (C) Effect of the triple L207A–W211A–F214A mutation on the avidity of [1–257]ArfGAP1 for Golgi-mix liposomes of defined size. Protein concentration: 0.75 μM; lipid concentration: 0.75 mM.
Figure 5
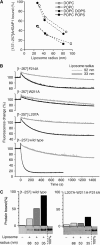
The central region of ArfGAP1 is highly susceptible to proteolysis in solution but is protected in the presence of small liposomes. ArfGAP1[1–257] was incubated with no liposomes, small DOPC:DOPS liposomes or large DOPC:DOPS liposomes. At time zero, chymotrypsin was added. The reaction was stopped at the indicated times and analysed by SDS–PAGE using sypro-orange staining (A) and by Western blot using an anti-polyhistidine tag antibody (B). The first line in the Western blot is a mixture of the indicated ArfGAP constructs (see Figure 3A), which were used as standards. Note that ArfGAP1[1–257] is slightly contaminated at time zero by a band that migrates between the first fragment that is generated by chymotrypsin (band 1) and the [1–196] standard. Similar results were observed with Golgi-mix liposomes. (C) Mass spectrometry analysis of the various fragments generated by chymotrypsin. (D) Schematic view of the proteolysis reaction.
Figure 6
The ALPS motif adopts a helical structure at the surface of highly curved membrane. (A) Far-UV CD spectra of full-length ArfGAP1 and of truncated constructs in the absence (black traces) or presence (red traces) of small DOPC/DOPS liposomes (hydrodynamic radius 24–30 nm). From left to right: ArfGAP1 (8 μM) with and without 4.6 mM liposomes (lipid/protein ratio=575); [1–196]ArfGAP1 (13 μM) with and without 3.225 mM liposomes (L/P=250); [137–257]ArfGAP1 (30 μM) with and without 7.5 mM liposomes (L/P=250); [192–257]ArfGAP (14 μM) with and without 3.125 mM liposomes (L/P=223). (B) Helical-wheel representation of the 199–223 sequence of rat ArfGAP1. The same colour code as Figure 1 was used. The yellow–purple gradation used for tyrosine illustrates the dual character of this residue (hydrophobic and hydroxylated). (C) Schematic view of the coupled folding binding process by which the ALPS motif recognizes curved membranes. Black diamonds represent hydrophobic residues. Chymotrypsin cleavage sites are shown in red. Note that ArfGAP1 interacts also with transmembrane proteins and with coatomer, which have been omitted in this scheme for clarity.
Similar articles
- Two lipid-packing sensor motifs contribute to the sensitivity of ArfGAP1 to membrane curvature.
Mesmin B, Drin G, Levi S, Rawet M, Cassel D, Bigay J, Antonny B. Mesmin B, et al. Biochemistry. 2007 Feb 20;46(7):1779-90. doi: 10.1021/bi062288w. Epub 2007 Jan 25. Biochemistry. 2007. PMID: 17253781 - Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature.
Bigay J, Gounon P, Robineau S, Antonny B. Bigay J, et al. Nature. 2003 Dec 4;426(6966):563-6. doi: 10.1038/nature02108. Nature. 2003. PMID: 14654841 - Identification of a second amphipathic lipid-packing sensor-like motif that contributes to Gcs1p function in the early endosome-to-TGN pathway.
Zendeh-boodi Z, Yamamoto T, Sakane H, Tanaka K. Zendeh-boodi Z, et al. J Biochem. 2013 Jun;153(6):573-87. doi: 10.1093/jb/mvt025. Epub 2013 Apr 5. J Biochem. 2013. PMID: 23564908 - Membrane curvature and the control of GTP hydrolysis in Arf1 during COPI vesicle formation.
Antonny B, Bigay J, Casella JF, Drin G, Mesmin B, Gounon P. Antonny B, et al. Biochem Soc Trans. 2005 Aug;33(Pt 4):619-22. doi: 10.1042/BST0330619. Biochem Soc Trans. 2005. PMID: 16042557 Review. - Amphipathic helices and membrane curvature.
Drin G, Antonny B. Drin G, et al. FEBS Lett. 2010 May 3;584(9):1840-7. doi: 10.1016/j.febslet.2009.10.022. Epub 2009 Oct 20. FEBS Lett. 2010. PMID: 19837069 Review.
Cited by
- A filter at the entrance of the Golgi that selects vesicles according to size and bulk lipid composition.
Magdeleine M, Gautier R, Gounon P, Barelli H, Vanni S, Antonny B. Magdeleine M, et al. Elife. 2016 Jul 26;5:e16988. doi: 10.7554/eLife.16988. Elife. 2016. PMID: 27458799 Free PMC article. - ArfGAP1 inhibits mTORC1 lysosomal localization and activation.
Meng D, Yang Q, Melick CH, Park BC, Hsieh TS, Curukovic A, Jeong MH, Zhang J, James NG, Jewell JL. Meng D, et al. EMBO J. 2021 Jun 15;40(12):e106412. doi: 10.15252/embj.2020106412. Epub 2021 May 14. EMBO J. 2021. PMID: 33988249 Free PMC article. - FisB mediates membrane fission during sporulation in Bacillus subtilis.
Doan T, Coleman J, Marquis KA, Meeske AJ, Burton BM, Karatekin E, Rudner DZ. Doan T, et al. Genes Dev. 2013 Feb 1;27(3):322-34. doi: 10.1101/gad.209049.112. Genes Dev. 2013. PMID: 23388828 Free PMC article. - Arf GAPs: gatekeepers of vesicle generation.
Spang A, Shiba Y, Randazzo PA. Spang A, et al. FEBS Lett. 2010 Jun 18;584(12):2646-51. doi: 10.1016/j.febslet.2010.04.005. Epub 2010 Apr 13. FEBS Lett. 2010. PMID: 20394747 Free PMC article. Review. - Curvature sensing as an emergent property of multiscale assembly of septins.
Shi W, Cannon KS, Curtis BN, Edelmaier C, Gladfelter AS, Nazockdast E. Shi W, et al. Proc Natl Acad Sci U S A. 2023 Feb 7;120(6):e2208253120. doi: 10.1073/pnas.2208253120. Epub 2023 Jan 30. Proc Natl Acad Sci U S A. 2023. PMID: 36716363 Free PMC article.
References
- Antonny B, Beraud-Dufour S, Chardin P, Chabre M (1997a) N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry 36: 4675–4684 - PubMed
- Antonny B, Huber I, Paris S, Chabre M, Cassel D (1997b) Activation of ADP-ribosylation factor 1 GTPase-activating protein by phosphatidylcholine-derived diacylglycerols. J Biol Chem 272: 30848–30851 - PubMed
- Antonny B, Madden D, Hamamoto S, Orci L, Schekman R (2001) Dynamics of the COPII coat with GTP and stable analogues. Nat Cell Biol 3: 531–537 - PubMed
- Antonny B, Schekman R (2001) ER export: public transportation by the COPII coach. Curr Opin Cell Biol 13: 438–443 - PubMed
- Aoe T, Huber I, Vasudevan C, Watkins SC, Romero G, Cassel D, Hsu VW (1999) The KDEL receptor regulates a GTPase-activating protein for ADP-ribosylation factor 1 by interacting with its non-catalytic domain. J Biol Chem 274: 20545–20549 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases