Gene movement by Helitron transposons contributes to the haplotype variability of maize - PubMed (original) (raw)
Gene movement by Helitron transposons contributes to the haplotype variability of maize
Jinsheng Lai et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Different maize inbred lines are polymorphic for the presence or absence of genic sequences at various allelic chromosomal locations. In the bz genomic region, located in 9S, sequences homologous to four different genes from rice and Arabidopsis are present in line McC but absent from line B73. It is shown here that this apparent intraspecific violation of genetic colinearity arises from the movement of genes or gene fragments by Helitrons, a recently discovered class of eukaryotic transposons. Two Helitrons, HelA and HelB, account for all of the genic differences distinguishing the two bz locus haplotypes. HelA is 5.9 kb long and contains sequences for three of the four genes found only in the McC bz genomic region. A nearly identical copy of HelA was isolated from a 5S chromosomal location in B73. Both the 9S and 5S sites appear to be polymorphic in maize, suggesting that these Helitrons have been active recently. Helitrons lack the strong predictive terminal features of other transposons, so the definition of their ends is greatly facilitated by the identification of their vacant sites in Helitron-minus lines. The ends of the 2.7-kb HelB Helitron were discerned from a comparison of the McC haplotype sequence with that of yet a third line, Mo17, because the HelB vacant site is deleted in B73. Maize Helitrons resemble rice Pack-MULEs in their ability to capture genes or gene fragments from several loci and move them around the genome, features that confer on them a potential role in gene evolution.
Figures
Fig. 1.
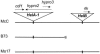
Organization of the ≈10-kb gene sequences from the McC bz haplotype that are missing from the B73 bz haplotype. (A) Structure of the McC region in 9S (from ref. 3). Predicted genes are shown as pentagons pointing in the direction of transcription. Exons are shown in bronze and introns in yellow. The proximal end is at 0 kb and the distal end at 10 kb. (B) Diagram of the
blastn
results, with the red bars representing the two contigs formed by overlapping GSS sequences from B73. (C) Structure of HelA and HelB, the two _Helitron_s that, together, account for all of the genes present in the bz genomic region of McC but not of B73. Helitron terminal sequences are in uppercase and the host target sequences are in lowercase. The stem-and-loop structure in each Helitron symbolizes the palindrome close to the 3′ end (not shown to scale; the palindrome does not overlap any gene). The location of the gene sequences derived from a reanalysis of genomic and cDNA sequences in the database is shown below each Helitron. The hypro3 gene was misannotated originally and does not span both _Helitron_s. (D) Exon–intron structure of the sequenced hypro2 cDNA clone from HelA2 (GenBank accession no. DQ000639). Exons are represented as thick straight lines and introns as thin angular lines.
Fig. 2.
Organization of a 50-kb sequence from the B73 chromosome 5 BAC b0511I12 (GenBank accession no. AC159612). A gene island is flanked on either side by retrotransposon clusters. The genes cdl and hypro2 are adjacent to each other at the left end of the gene island (marked by the dotted-perimeter box). Part of the predicted hypro3 gene is nested within hypro2. The other predicted genes in the gene island are: 3, hypothetical protein; 4, esterase; 5, aldehyde oxidase; 6, β-1,3-glucanase; 7, replication factor; and 8, hypothetical protein.
Fig. 3.
Termini of the maize _Helitron_s HelA-1 and HelB from McC 9S, _HelA_-2 from B73 5S, the Helitron insertions in mutants _sh2_-7527 (13) and ba1-Ref (14, 16), and the rice Helitron2_ OS (12). Helitron sequences are in uppercase letters and the invariant host nucleotides where the _Helitron_s insert are in lowercase letters. Conserved nucleotides at the 5′ and 3′ termini are in bold uppercase letters and the inverted repeats at the 3′ termini are underlined.
Fig. 4.
The Helitron structure of bz haplotypes McC, B73, and Mo17. The _Helitron_s are shown as downward-pointing triangles, and the corresponding vacant sites in B73 and Mo17 are represented by thick short vertical lines. The vacant site for HelB is missing in B73.
Fig. 5.
Diagram of the origin of an McC-type haplotype from a Mo17-type progenitor haplotype by the transposition of _Helitron_s. _Helitron_s HelA and HelB are represented as thick gray lines in the donor chromosomes. Putative RC-transposition intermediates are shown as circles inserting into the vacant target AT dinucleotides of a Mo17-like haplotype (shown here, for simplicity, in the same 5′–3′ orientation). The resulting transposition product would be an McC-like haplotype.
Comment in
- Helitrons contribute to the lack of gene colinearity observed in modern maize inbreds.
Lal SK, Hannah LC. Lal SK, et al. Proc Natl Acad Sci U S A. 2005 Jul 19;102(29):9993-4. doi: 10.1073/pnas.0504713102. Epub 2005 Jul 11. Proc Natl Acad Sci U S A. 2005. PMID: 16009929 Free PMC article. No abstract available.
Similar articles
- Computational prediction and molecular confirmation of Helitron transposons in the maize genome.
Du C, Caronna J, He L, Dooner HK. Du C, et al. BMC Genomics. 2008 Jan 28;9:51. doi: 10.1186/1471-2164-9-51. BMC Genomics. 2008. PMID: 18226261 Free PMC article. - The polychromatic Helitron landscape of the maize genome.
Du C, Fefelova N, Caronna J, He L, Dooner HK. Du C, et al. Proc Natl Acad Sci U S A. 2009 Nov 24;106(47):19916-21. doi: 10.1073/pnas.0904742106. Epub 2009 Nov 19. Proc Natl Acad Sci U S A. 2009. PMID: 19926866 Free PMC article. - Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus.
Wang Q, Dooner HK. Wang Q, et al. Proc Natl Acad Sci U S A. 2006 Nov 21;103(47):17644-9. doi: 10.1073/pnas.0603080103. Epub 2006 Nov 13. Proc Natl Acad Sci U S A. 2006. PMID: 17101975 Free PMC article. - Helitrons on a roll: eukaryotic rolling-circle transposons.
Kapitonov VV, Jurka J. Kapitonov VV, et al. Trends Genet. 2007 Oct;23(10):521-9. doi: 10.1016/j.tig.2007.08.004. Epub 2007 Sep 11. Trends Genet. 2007. PMID: 17850916 Review. - Helitrons: genomic parasites that generate developmental novelties.
Barro-Trastoy D, Köhler C. Barro-Trastoy D, et al. Trends Genet. 2024 May;40(5):437-448. doi: 10.1016/j.tig.2024.02.002. Epub 2024 Feb 29. Trends Genet. 2024. PMID: 38429198 Review.
Cited by
- Highly asymmetric rice genomes.
Ding J, Araki H, Wang Q, Zhang P, Yang S, Chen JQ, Tian D. Ding J, et al. BMC Genomics. 2007 Jun 8;8:154. doi: 10.1186/1471-2164-8-154. BMC Genomics. 2007. PMID: 17555605 Free PMC article. - Cisgenic plants are similar to traditionally bred plants: international regulations for genetically modified organisms should be altered to exempt cisgenesis.
Schouten HJ, Krens FA, Jacobsen E. Schouten HJ, et al. EMBO Rep. 2006 Aug;7(8):750-3. doi: 10.1038/sj.embor.7400769. EMBO Rep. 2006. PMID: 16880817 Free PMC article. No abstract available. - Helitrons contribute to the lack of gene colinearity observed in modern maize inbreds.
Lal SK, Hannah LC. Lal SK, et al. Proc Natl Acad Sci U S A. 2005 Jul 19;102(29):9993-4. doi: 10.1073/pnas.0504713102. Epub 2005 Jul 11. Proc Natl Acad Sci U S A. 2005. PMID: 16009929 Free PMC article. No abstract available. - Miniature inverted-repeat transposable elements (MITEs), derived insertional polymorphism as a tool of marker systems for molecular plant breeding.
Venkatesh, Nandini B. Venkatesh, et al. Mol Biol Rep. 2020 Apr;47(4):3155-3167. doi: 10.1007/s11033-020-05365-y. Epub 2020 Mar 11. Mol Biol Rep. 2020. PMID: 32162128 Review.
References
- SanMiguel, P. & Bennetzen, J. L. (1998) Ann. Bot. (London) 82, 37–44.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources